Structure of the Capsid Size-Determining Scaffold of “Satellite” Bacteriophage P4
Abstract
:1. Introduction
2. Materials and Methods
2.1. Production of P4 Procapsids
2.2. Cryo-Electron Microscopy
2.3. Three-Dimensional Reconstruction and Model Building
2.4. Model Building and Refinement
3. Results
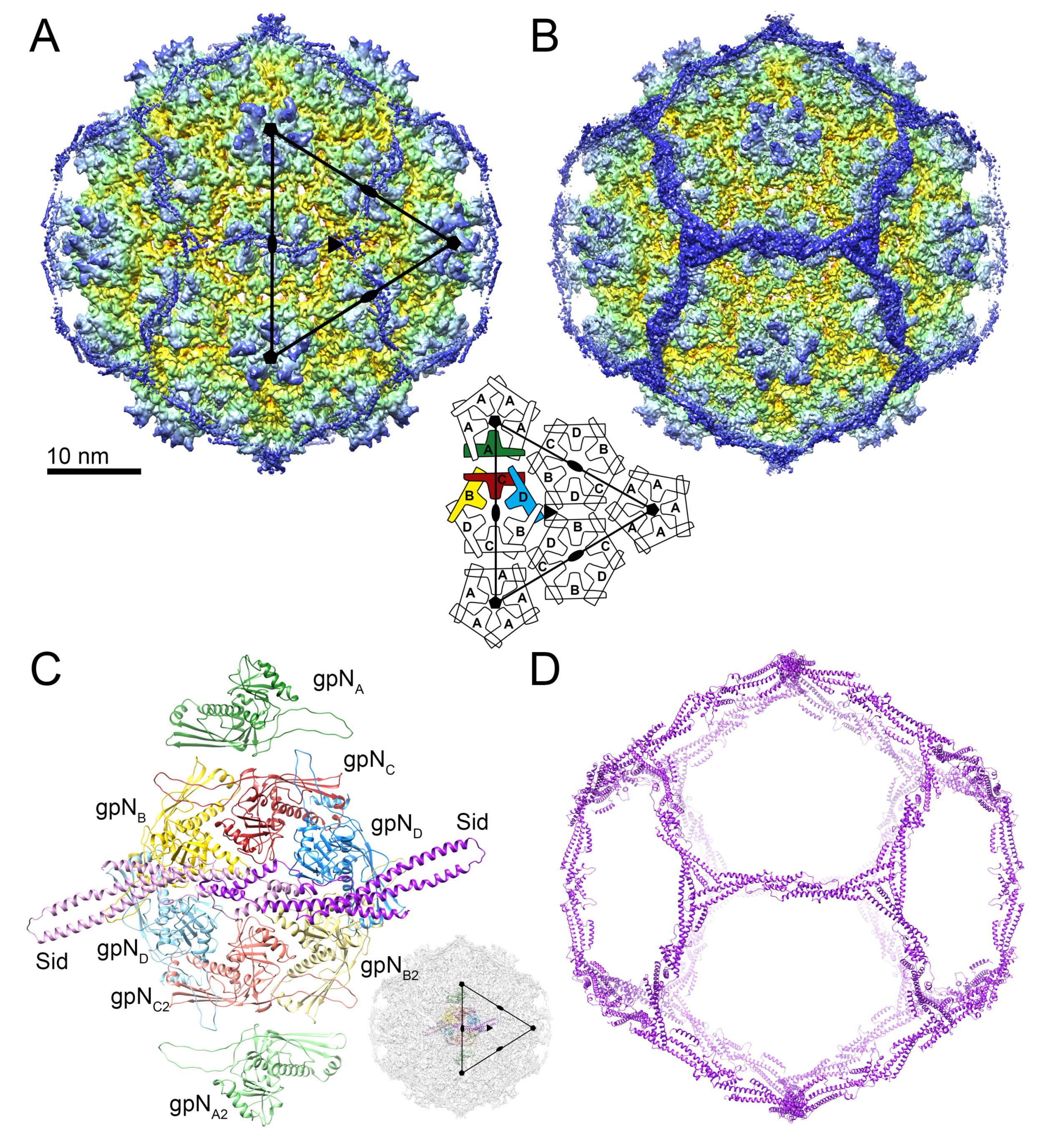
3.1. Structure Determination
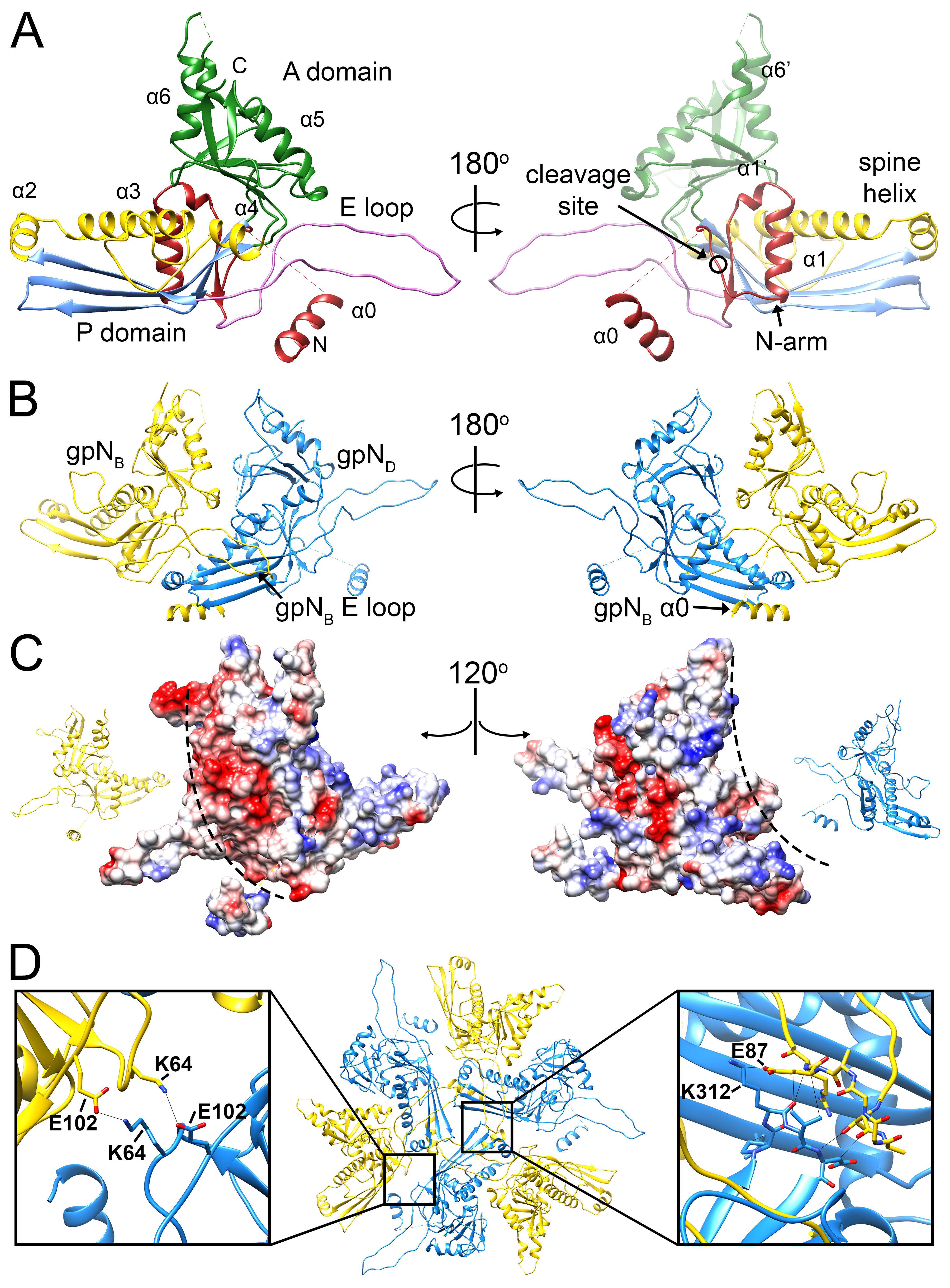
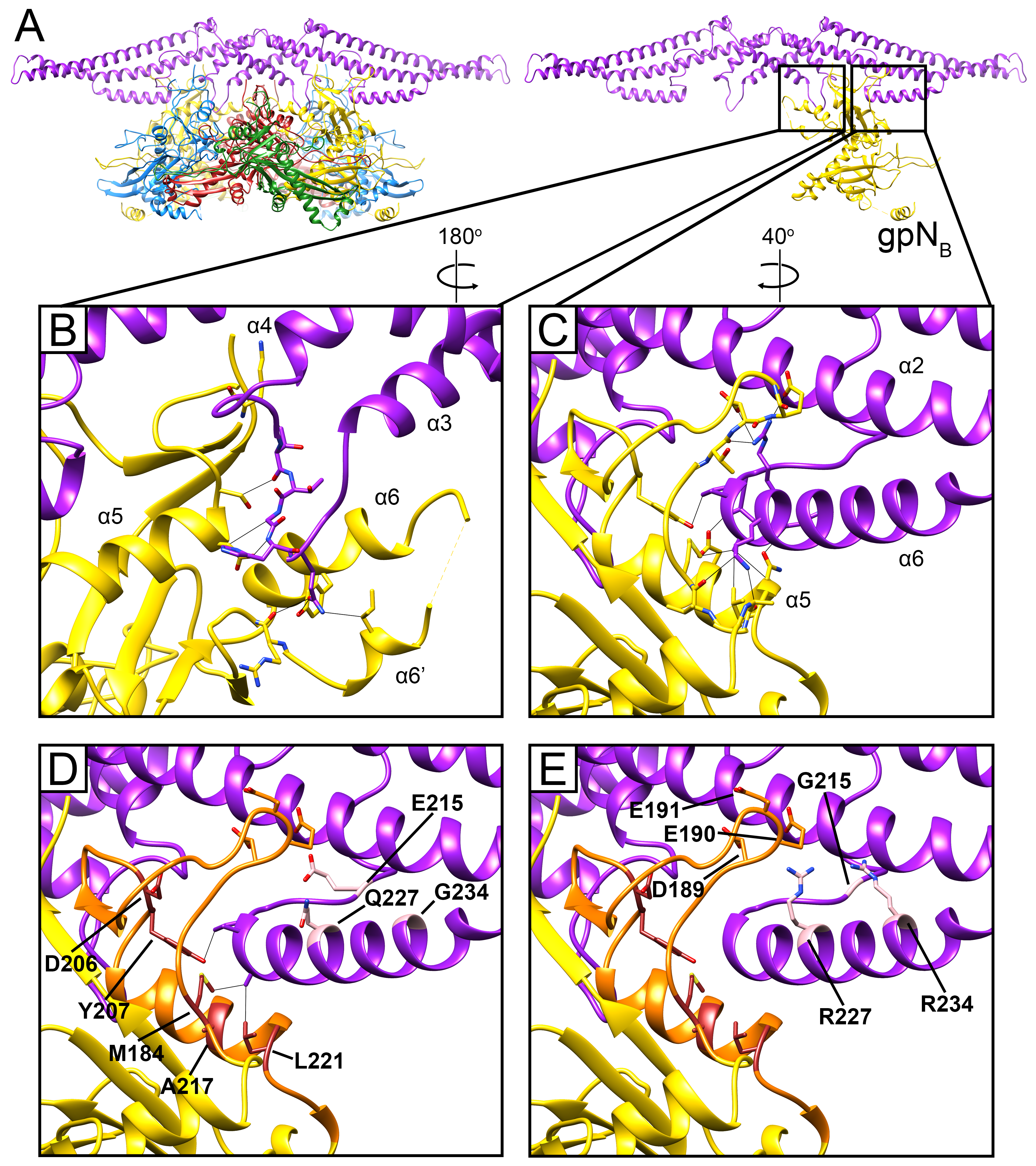
3.2. The gpN Capsid Protein
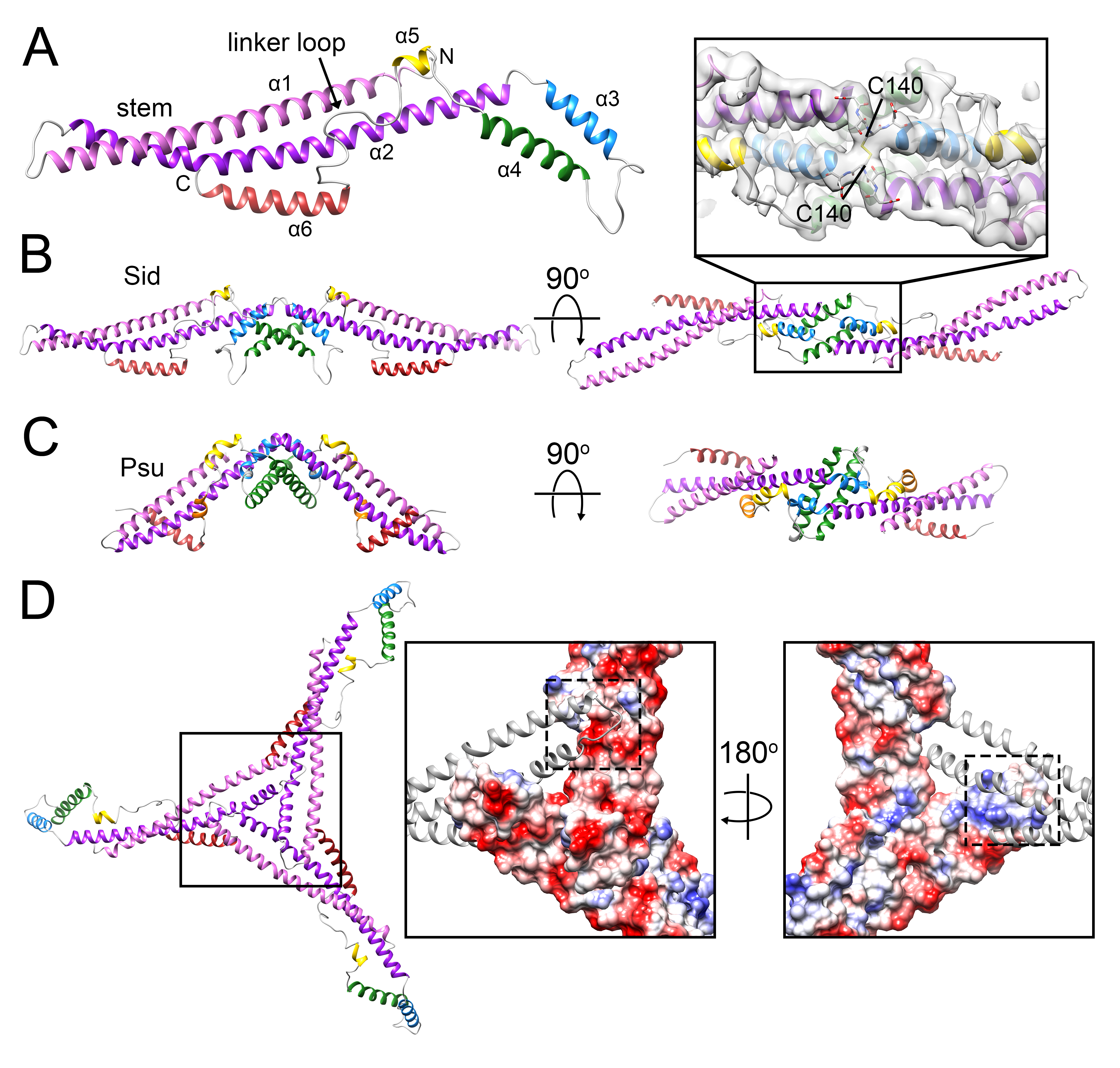
3.3. The External Sid Scaffold
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Fane, B.A.; Brentlinger, K.L.; Burch, A.D.; Chen, M.; Hafenstein, S.; Moore, E.; Novak, C.R.; Uchiyama, A. The Microviridae. In The Bacteriophages; Calendar, R., Ed.; Oxford University Press: New York, NY, USA, 2006; pp. 129–145. [Google Scholar]
- Dokland, T.; McKenna, R.; Ilag, L.L.; Bowman, B.R.; Incardona, N.L.; Fane, B.A.; Rossmann, M.G. Structure of a viral procapsid with molecular scaffolding. Nature 1997, 389, 308–313. [Google Scholar] [CrossRef] [PubMed]
- Ackermann, H.W. Tailed bacteriophages: The order caudovirales. Adv. Virus Res. 1998, 51, 135–201. [Google Scholar] [PubMed]
- Marvik, O.J.; Dokland, T.; Nøkling, R.H.; Jacobsen, E.; Larsen, T.; Lindqvist, B.H. The capsid size-determining protein Sid forms an external scaffold on phage P4 procapsids. J. Mol. Biol. 1995, 251, 59–75. [Google Scholar] [CrossRef] [PubMed]
- Christie, G.E.; Calendar, R. Bacteriophage P2. Bacteriophage 2016, 6, e1145782. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dokland, T.; Lindqvist, B.H.; Fuller, S.D. Image reconstruction from cryo-electron micrographs reveals the morphopoietic mechanism in the P2-P4 bacteriophage system. EMBO J. 1992, 11, 839–846. [Google Scholar] [CrossRef]
- Chang, J.R.; Poliakov, A.; Prevelige, P.E.; Mobley, J.A.; Dokland, T. Incorporation of scaffolding protein gpO in bacteriophages P2 and P4. Virology 2008, 370, 352–361. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nilsson, A.S.; Karlsson, J.L.; Haggård-Ljungquist, E. Site-specific recombination links the evolution of P2-like coliphages and pathogenic enterobacteria. Mol. Biol. Evol. 2004, 21, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Dearborn, A.D.; Laurinmaki, P.; Chandramouli, P.; Rodenburg, C.M.; Wang, S.; Butcher, S.J.; Dokland, T. Structure and size determination of bacteriophage P2 and P4 procapsids: Function of size responsiveness mutations. J. Struct. Biol. 2012, 178, 215–224. [Google Scholar] [CrossRef] [Green Version]
- Rishovd, S.; Marvik, O.J.; Jacobsen, E.; Lindqvist, B.H. Bacteriophage P2 and P4 morphogenesis: Identification and characterization of the portal protein. Virology 1994, 200, 744–751. [Google Scholar] [CrossRef]
- Rishovd, S.; Holzenburg, A.; Johansen, B.V.; Lindqvist, B.H. Bacteriophage P2 and P4 morphogenesis: Structure and function of the connector. Virology 1998, 245, 11–17. [Google Scholar] [CrossRef] [Green Version]
- Chang, J.R.; Spilman, M.S.; Rodenburg, C.M.; Dokland, T. Functional domains of the bacteriophage P2 scaffolding protein: Identification of residues involved in assembly and protease activity. Virology 2009, 384, 144–150. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rishovd, S.; Lindqvist, B.H. Bacteriophage P2 and P4 morphogenesis: Protein processing and capsid size determination. Virology 1992, 187, 548–554. [Google Scholar] [CrossRef]
- Bowden, D.W.; Modrich, P. In vitro maturation of circular bacteriophage P2 DNA. J. Biol. Chem. 1985, 260, 6999–7007. [Google Scholar] [PubMed]
- Christie, G.E.; Dokland, T. Pirates of the Caudovirales. Virology 2012, 434, 210–221. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dokland, T. Molecular Piracy: Redirection of Bacteriophage Capsid Assembly by Mobile Genetic Elements. Viruses 2019, 11, 1003. [Google Scholar] [CrossRef] [Green Version]
- Christie, G.E.; Calendar, R. Interactions between satellite bacteriophage P4 and its helpers. Annu. Rev. Genet. 1990, 24, 465–490. [Google Scholar] [CrossRef]
- Halling, C.; Calendar, R. Bacteriophage P2 ogr and P4 delta genes act independently and are essential for P4 multiplication. J. Bacteriol. 1990, 172, 3549–3558. [Google Scholar] [CrossRef] [Green Version]
- Sunshine, M.; Six, E.; Barrett, K.; Calendar, R. Relief of P2 bacteriophage amber mutant polarity by the satellite bacteriophage P4. J. Mol. Biol. 1976, 106, 673–682. [Google Scholar] [CrossRef]
- Dokland, T.; Isaksen, M.L.; Fuller, S.D.; Lindqvist, B.H. Capsid Localization of the Bacteriophage P4 Psu Protein. Virology 1993, 194, 682–687. [Google Scholar] [CrossRef]
- Isaksen, M.L.; Dokland, T.; Lindqvist, B.H. Characterization of the Capsid Associating Activity of Bacteriophage P4′s Psu Protein. Virology 1993, 194, 674–681. [Google Scholar] [CrossRef]
- Pani, B.; Ranjan, A.; Sen, R. Interaction Surface of Bacteriophage P4 Protein Psu Required for Complex Formation with the Transcription Terminator Rho. J. Mol. Biol. 2009, 389, 647–660. [Google Scholar] [CrossRef] [PubMed]
- Ranjan, A.; Sharma, S.; Banerjee, R.; Sen, U.; Sen, R. Structural and mechanistic basis of anti-termination of Rho-dependent transcription termination by bacteriophage P4 capsid protein Psu. Nucleic Acids Res. 2013, 41, 6839–6856. [Google Scholar] [CrossRef] [PubMed]
- Shore, D.; Dehò, G.; Tsipis, J.; Goldstein, R. Determination of capsid size by satellite bacteriophage P4. Proc. Natl. Acad. Sci. USA 1978, 75, 400–404. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, T.; Renberg, S.K.; Haggård-Ljungquist, E. Derepression of prophage P2 by satellite phage P4: Cloning of the P4 epsilon gene and identification of its product. J. Virol. 1997, 71, 4502–4508. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Six, E.W.; Sunshine, M.G.; Williams, J.; Haggård-Ljungquist, E.; Lindqvist, B.H. Morphopoietic switch mutations of bacteriophage P2. Virology 1991, 182, 34–46. [Google Scholar] [CrossRef]
- Kim, K.-J.; Sunshine, M.G.; Lindqvist, B.H.; Six, E.W. Capsid Size Determination in the P2–P4 Bacteriophage System: Suppression of sir Mutations in P2′s Capsid Gene N by Supersid Mutations in P4′s External Scaffold Gene sid. Virology 2001, 283, 49–58. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zivanov, J.; Nakane, T.; O Forsberg, B.; Kimanius, D.; Hagen, W.J.H.; Lindahl, E.; Scheres, S.H.W. New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife 2018, 7, 42166. [Google Scholar] [CrossRef]
- Zheng, S.Q.; Palovcak, E.; Armache, J.-P.; Verba, K.A.; Cheng, Y.; Agard, D.A. MotionCor2: Anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 2017, 14, 331–332. [Google Scholar] [CrossRef] [Green Version]
- Zhang, K. Gctf: Real-time CTF determination and correction. J. Struct. Biol. 2016, 193, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Scheres, S.H.W. Processing of Structurally Heterogeneous Cryo-EM Data in RELION. Methods Enzymol. 2016, 579, 125–157. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Afonne, P.V.; Klaholz, B.P.; Moriarty, N.W.; Poon, B.K.; Sobolev, O.V.; Terwilliger, T.C.; Adams, P.D.; Urzhumtsev, A. New tools for the analysis and validation of cryo-EM maps and atomic models. Acta Crystallogr. Sect. D Struct. Biol. 2018, 74, 814–840. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, J.; Yan, R.; Roy, A.; Xu, N.; Poisson, J.; Zhang, Y. The I-TASSER Suite: Protein structure and function prediction. Nat. Methods 2014, 12, 7–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Emsley, P.; Lohkamp, B.; Scott, W.G.; Cowtan, K. Features and development of Coot. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010, 66, 486–501. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, V.B.; Arendall, W.B.; Headd, J.J.; Keedy, D.A.; Immormino, R.M.; Kapral, G.J.; Murray, L.W.; Richardson, J.S.; Richardson, D.C. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr. Sect. D Biol. Crystallogr. 2009, 66, 12–21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barad, B.A.; Echols, N.; Wang, R.Y.-R.; Cheng, Y.; DiMaio, F.; Adams, P.D.; Fraser, J.S. EMRinger: Side chain–directed model and map validation for 3D cryo-electron microscopy. Nat. Methods 2015, 12, 943–946. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duda, R.L.; Teschke, C.M. The amazing HK97 fold: Versatile results of modest differences. Curr. Opin. Virol. 2019, 36, 9–16. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.-H.; Baker, M.L.; Hryc, C.F.; DiMaio, F.; Jakana, J.; Wu, W.; Dougherty, M.; Haase-Pettingell, C.; Schmid, M.F.; Jiang, W.; et al. Structural basis for scaffolding-mediated assembly and maturation of a dsDNA virus. Proc. Natl. Acad. Sci. USA 2011, 108, 1355–1360. [Google Scholar] [CrossRef] [Green Version]
- Dearborn, A.D.; Wall, E.A.; Kizziah, J.L.; Klenow, L.; Parker, L.K.; Manning, K.A.; Spilman, M.S.; Spear, J.M.; Christie, G.E.; Dokland, T. Competing scaffolding proteins determine capsid size during mobilization of Staphylococcus aureus pathogenicity islands. eLife 2017, 6, 30822. [Google Scholar] [CrossRef]
- Helgstrand, C.; Wikoff, W.R.; Duda, R.L.; Hendrix, R.W.; E Johnson, J.; Liljas, L. The Refined Structure of a Protein Catenane: The HK97 Bacteriophage Capsid at 3.44 Å Resolution. J. Mol. Biol. 2003, 334, 885–899. [Google Scholar] [CrossRef]
- Lander, G.C.; Evilevitch, A.; Jeembaeva, M.; Potter, C.S.; Carragher, B.; Johnson, J.E. Bacteriophage Lambda Stabilization by Auxiliary Protein gpD: Timing, Location, and Mechanism of Attachment Determined by Cryo-EM. Structure 2008, 16, 1399–1406. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Landeta, C.; Boyd, D.; Beckwith, J. Disulfide bond formation in prokaryotes. Nat. Microbiol. 2018, 3, 270–280. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Palasingam, P.; Nøkling, R.H.; Lindqvist, B.H.; Dokland, T. In Vitro Assembly of Bacteriophage P4 Procapsids from Purified Capsid and Scaffolding Proteins. Virology 2000, 275, 133–144. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Banerjee, R.; Nath, S.; Ranjan, A.; Khamrui, S.; Pani, B.; Sen, R.; Sen, U. The First Structure of Polarity Suppression Protein, Psu from Enterobacteria Phage P4, Reveals a Novel Fold and a Knotted Dimer. J. Biol. Chem. 2012, 287, 44667–44675. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- DeDeo, C.L.; Cingolani, G.; Teschke, C.M. Portal Protein: The Orchestrator of Capsid Assembly for the dsDNA Tailed Bacteriophages and Herpesviruses. Annu. Rev. Virol. 2019, 6, 141–160. [Google Scholar] [CrossRef] [PubMed]
- Cardarelli, L.; Pell, L.G.; Neudecker, P.; Pirani, N.; Liu, A.; Baker, L.A.; Rubinstein, J.L.; Maxwell, K.L.; Davidson, A.R. Phages have adapted the same protein fold to fulfill multiple functions in virion assembly. Proc. Natl. Acad. Sci. USA 2010, 107, 14384–14389. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goulet, A.; Spinelli, S.; Mahony, J.; Cambillau, C. Conserved and Diverse Traits of Adhesion Devices from Siphoviridae Recognizing Proteinaceous or Saccharidic Receptors. Viruses 2020, 12, 512. [Google Scholar] [CrossRef]
- Morais, M.C.; Kanamaru, S.; O Badasso, M.; Koti, J.S.; Owen, B.A.L.; McMurray, C.T.; Anderson, D.L.; Rossmann, M.G. Bacteriophage φ29 scaffolding protein gp7 before and after prohead assembly. Nat. Struct. Mol. Biol. 2003, 10, 572–576. [Google Scholar] [CrossRef]
- Sun, Y.; Parker, M.H.; Weigele, P.; Casjens, S.; Prevelige, P.E., Jr.; Krishna, N. Structure of the coat protein-binding domain of the scaffolding protein from a double-stranded DNA virus11Edited by M. Summers. J. Mol. Biol. 2000, 297, 1195–1202. [Google Scholar] [CrossRef] [Green Version]
- Dearborn, A.D.; Spilman, M.S.; Damle, P.K.; Chang, J.R.; Monroe, E.B.; Saad, J.S.; Christie, G.E.; Dokland, T. The Staphylococcus aureus Pathogenicity Island 1 Protein gp6 Functions as an Internal Scaffold during Capsid Size Determination. J. Mol. Biol. 2011, 412, 710–722. [Google Scholar] [CrossRef] [Green Version]
- Morais, M.C.; Fisher, M.; Kanamaru, S.; Przybyla, L.; Burgner, J.; Fane, B.A.; Rossmann, M.G. Conformational Switching by the Scaffolding Protein D Directs the Assembly of Bacteriophage φX174. Mol. Cell 2004, 15, 991–997. [Google Scholar] [CrossRef] [PubMed]
- Henne, W.M.; Kent, H.M.; Ford, M.G.; Hegde, B.G.; Daumke, O.; Butler, P.J.G.; Mittal, R.; Langen, R.; Evans, P.R.; McMahon, H.T. Structure and Analysis of FCHo2 F-BAR Domain: A Dimerizing and Membrane Recruitment Module that Effects Membrane Curvature. Structure 2007, 15, 839–852. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zimmermann, L.; Stephens, A.; Nam, S.-Z.; Rau, D.; Kübler, J.; Lozajic, M.; Gabler, F.; Söding, J.; Lupas, A.N.; Alva, V. A Completely Reimplemented MPI Bioinformatics Toolkit with a New HHpred Server at its Core. J. Mol. Biol. 2018, 430, 2237–2243. [Google Scholar] [CrossRef] [PubMed]
- Dokland, T. Freedom and restraint: Themes in virus capsid assembly. Structure 2000, 8, R157–R162. [Google Scholar] [CrossRef] [Green Version]
- Zlotnick, A. Theoretical aspects of virus capsid assembly. J. Mol. Recognit. 2005, 18, 479–490. [Google Scholar] [CrossRef]
- Fu, C.-Y.; Uetrecht, C.; Kang, S.; Morais, M.C.; Heck, A.; Walter, M.R.; Prevelige, P.E. A Docking Model Based on Mass Spectrometric and Biochemical Data Describes Phage Packaging Motor Incorporation. Mol. Cell. Proteom. 2010, 9, 1764–1773. [Google Scholar] [CrossRef] [Green Version]


© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kizziah, J.L.; Rodenburg, C.M.; Dokland, T. Structure of the Capsid Size-Determining Scaffold of “Satellite” Bacteriophage P4. Viruses 2020, 12, 953. https://0-doi-org.brum.beds.ac.uk/10.3390/v12090953
Kizziah JL, Rodenburg CM, Dokland T. Structure of the Capsid Size-Determining Scaffold of “Satellite” Bacteriophage P4. Viruses. 2020; 12(9):953. https://0-doi-org.brum.beds.ac.uk/10.3390/v12090953
Chicago/Turabian StyleKizziah, James L., Cynthia M. Rodenburg, and Terje Dokland. 2020. "Structure of the Capsid Size-Determining Scaffold of “Satellite” Bacteriophage P4" Viruses 12, no. 9: 953. https://0-doi-org.brum.beds.ac.uk/10.3390/v12090953




