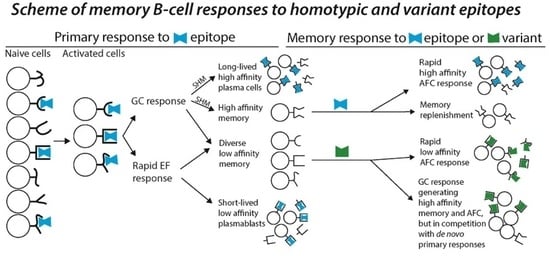
B-Cell Memory Responses to Variant Viral Antigens
Abstract
:1. B-Cell Memory and Its Cross-Reactivity
2. Viral Point Mutations and B-Cell Memory Responses
3. SARS-CoV-2 Variants and Antibody Immunity
4. Concluding Remarks
Author Contributions
Funding
Conflicts of Interest
References
- Weisel, F.; Shlomchik, M. Memory B cells of mice and humans. Annu. Rev. Immunol. 2017, 35, 255–284. [Google Scholar] [CrossRef]
- Kurosaki, T.; Kometani, K.; Ise, W. Memory B cells. Nat. Rev. Immunol. 2015, 15, 149–159. [Google Scholar] [CrossRef] [PubMed]
- Seifert, M.; Küppers, R. Human memory B cells. Leukemia 2016, 30, 2283–2292. [Google Scholar] [CrossRef]
- Phan, T.G.; Tangye, S.G. Memory B cells: Total recall. Curr. Opin. Immunol. 2017, 45, 132–140. [Google Scholar] [CrossRef]
- Akkaya, M.; Kihyuck, K.; Pierce, S.K. B cell memory: Building two walls of protection against pathogens. Nat. Rev. Immunol. 2020, 20, 229–238. [Google Scholar] [CrossRef]
- Havenar-Daughton, C.; Abbott, R.K.; Schief, W.R.; Crotty, S. When designing vaccines, consider the starting material: The human B cell repertoire. Curr. Opin. Immunol. 2018, 53, 209–216. [Google Scholar] [CrossRef] [PubMed]
- MacLennan, I.C.; Gulbranson-Judge, A.; Toellner, K.M.; Casamayor-Palleja, M.; Sze, D.M.; Chan, E.Y.; Luther, S.A.; Orbea, H.A. The changing preference of T and B cells for partners as T-dependent antibody responses develop. Immunol. Rev. 1997, 156, 53–66. [Google Scholar] [CrossRef] [PubMed]
- Seikiguchi, J.; Alt, F.J.; Oettinger, M. The mechanism of VDJ recombination. In The Molecular Biology of B Cells, 2nd ed.; Elsevier: Amsterdam, The Netherlands, 2004; pp. 61–82. [Google Scholar]
- Cyster, J.G. B cell follicles and antigen encounters of the third kind. Nat. Immunol. 2010, 11, 989–996. [Google Scholar] [CrossRef] [PubMed]
- Allen, C.D.; Cyster, J.G. Follicular dendritic cell networks of primary follicles and germinal centers: Phenotype and function. Semin. Immunol. 2008, 20, 14–25. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heesters, B.A.; Myers, R.C.; Carroll, M.C. Follicular dendritic cells: Dynamic antigen libraries. Nat. Rev. Immunol. 2014, 14, 495–504. [Google Scholar] [CrossRef]
- Schulz, O.; Hammerschmidt, S.I.; Moschovakis, G.L.; Förster, R. Chemokines and Chemokine Receptors in Lymphoid Tissue Dynamics. Annu. Rev. Immunol. 2016, 34, 203–242. [Google Scholar] [CrossRef]
- Rock, K.L.; Benacerraf, B.; Abbas, A.K. Antigen presentation by hapten-specific B lymphocytes. I. Role of surface immunoglobulin receptors. J. Exp. Med. 1984, 160, 1102–1113. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lanzavecchia, A. Antigen-specific interaction between T and B cells. Nature 1985, 314, 537–539. [Google Scholar] [CrossRef] [PubMed]
- Cyster, J.G.; Dang, E.V.; Reboldi, A.; Yi, T. 25-Hydroxycholesterols in innate and adaptive immunity. Nat. Rev. Immunol. 2014, 14, 731–743. [Google Scholar] [CrossRef] [PubMed]
- Garside, P.; Ingulli, E.; Merica, R.R.; Johnson, J.G.; Noelle, R.J.; Jenkins, M.K. Visualization of specific B and T lymphocyte interactions in the lymph node. Science 1998, 281, 96–99. [Google Scholar] [CrossRef]
- Kaji, T.; Ishige, A.; Hikida, M.; Tako, J.; Hijikata, A.; Kubo, M.; Nagashima, T.; Takahashi, Y.; Kurosaki, T.; Okada, M.; et al. Distinct cellular pathways select germline-encoded and somatically mutated antibodies into immunological memory. J. Exp. Med. 2012, 209, 2079–2097. [Google Scholar] [CrossRef] [Green Version]
- Weisel, F.J.; Zuccarino-Catania, G.V.; Chikina, M.; Shlomchik, M.J. A temporal switch in the germinal center determines differential output of memory B and plasma cells. Immunity 2016, 44, 116–130. [Google Scholar] [CrossRef] [Green Version]
- Elsner, R.A.; Shlomchik, M.J. Germinal center and extrafollicular B cell responses in vaccination, immunity, and autoimmunity. Immunity 2020, 53, 1136–1150. [Google Scholar] [CrossRef]
- Viant, C.; Weymar, G.H.; Escolano, A.; Chen, S.; Hartweger, H.; Cipolla, M.; Gazumyan, A.; Nussenzweig, M.C. Antibody affinity shapes the choice between memory and germinal center B cell fates. Cell 2020, 183, 1298–1311. [Google Scholar] [CrossRef]
- Bannard, O.; Cyster, J.G. Germinal centers: Programmed for affinity maturation and antibody diversification. Curr. Opin. Immunol. 2017, 45, 30–31. [Google Scholar] [CrossRef]
- Cyster, J.G.; Allen, C.D.C. B cell responses: Cell interaction dynamics and decisions. Cell 2019, 177, 524–540. [Google Scholar] [CrossRef] [Green Version]
- Batista, F.D.; Neuberger, M.S. Affinity dependence of the B cell response to antigen: A threshold, a ceiling, and the importance of off-rate. Immunity 1998, 8, 751–759. [Google Scholar] [CrossRef] [Green Version]
- Dogan, I.; Bertocci, B.; Vilmont, V.; Delbos, F.; Mégret, J.; Storck, S.; Reynaud, C.-A.; Weill, J.-C. Multiple layers of B cell memory with different effector functions. Nat. Immunol. 2009, 10, 1292–1299. [Google Scholar] [CrossRef]
- Burton, B.R.; Tennant, R.K.; Love, J.; Titball, R.W.; Wraith, D.C.; White, H.N. Variant proteins stimulate more IgM+ GC B-cells revealing a mechanism of cross-reactive recognition by antibody memory. eLife 2018. [Google Scholar] [CrossRef] [PubMed]
- Mesin, L.; Schiepers, A.; Ersching, J.; Barbulescu, A.; Cavazzoni, C.B.; Angelini, A.; Okada, T.; Kurosaki, T.; Victora, G.D. Restricted clonality and limited germinal center reentry characterize memory B cell reactivation by boosting. Cell 2020, 9, 92–106. [Google Scholar] [CrossRef] [Green Version]
- Wong, R.; Belk, J.A.; Govero, J.; Uhrlaub, J.L.; Reinartz, D.; Zhao, H.; Errico, J.M.; D’Souza, L.; Ripperger, T.J.; Nikolich-Zugich, J.; et al. Affinity-restricted memory B cells dominate recall responses to heterologous flaviviruses. Immunity 2020, 53, 1078–1094. [Google Scholar] [CrossRef]
- Dejnirattisai, W.; Jumnainsong, A.; Onsirisakul, N.; Fitton, P.; Vasanawathana, S.; Limpitikul, W.; Puttikhunt, C.; Edwards, C.; Duangchinda, T.; Supasa, S. Enhancing cross-reactive anti-prM dominates the human antibody response in dengue infection. Science 2010, 328, 745–748. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McHeyzer-Williams, M.G.; Nossal, G.J.; Lalor, P.A. Molecular characterization of single memory B cells. Nature 1991, 350, 502–505. [Google Scholar] [CrossRef] [PubMed]
- Rajewsky, K. Clonal selection and learning in the antibody system. Nature 1996, 381, 751–758. [Google Scholar] [CrossRef]
- White, H.; Gray, D. Analysis of immunoglobulin (Ig) isotype diversity and IgM/D memory in the response to phenyl-oxazolone. J. Exp. Med. 2000, 191, 2209–2220. [Google Scholar] [CrossRef] [PubMed]
- Pape, K.A.; Taylor, J.J.; Maul, R.W.; Gearhart, P.J.; Jenkins, M.K. Different B cell populations mediate early and late memory during an endogenous immune response. Science 2011, 331, 1203–1207. [Google Scholar] [CrossRef] [Green Version]
- Zuccarino-Catania, G.V.; Sadanand, S.; Weisel, F.J.; Tomayko, M.M.; Meng, H.; Kleinstein, S.H.; Good-Jacobson, K.L.; Shlomchik, M.J. CD80 and PD-L2 define functionally distinct memory B cell subsets that are independent of antibody isotype. Nat. Immunol. 2014, 15, 631–637. [Google Scholar] [CrossRef]
- Dougan, D.A.; Malby, R.L.; Gruen, L.C.; Kortt, A.A.; Hudson, P.J. Effects of substitutions in the binding surface of an antibody on antigen affinity. Protein Eng. 1998, 11, 65–74. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Doud, M.B.; Lee, J.M.; Bloom, J.D. How single mutations affect viral escape from broad and narrow antibodies to H1 influenza hemagglutinin. Nat. Commun. 2018, 9, 1–2. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, K.Y.; Rijal, P.; Schimanski, L.; Powell, T.J.; Lin, T.Y.; McCauley, J.W.; Daniels, R.S.; Townsend, A.R. Focused antibody response to influenza linked to antigenic drift. J. Clin. Investig. 2015, 125, 2631–2645. [Google Scholar] [CrossRef] [PubMed]
- Gromowski, G.D.; Barrett, A.D. Characterization of an antigenic site that contains a dominant, type-specific neutralization determinant on the envelope protein domain III (ED3) of dengue 2 virus. Virology 2007, 366, 349–360. [Google Scholar] [CrossRef] [Green Version]
- Sanjuán, R.; Domingo-Calap, P. Mechanisms of viral mutation. Cell. Mol. Life Sci. 2016, 73, 4433–4448. [Google Scholar] [CrossRef] [Green Version]
- Foote, J.; Milstein, C. Conformational isomerism and the diversity of antibodies. Proc. Natl. Acad. Sci. USA 1994, 91, 10370–10374. [Google Scholar] [CrossRef] [Green Version]
- Wedemayer, G.J.; Patten, P.A.; Wang, L.H.; Schultz, P.G.; Stevens, R.C. Structural insights into the evolution of an antibody combining site. Science 1997, 276, 1665–1669. [Google Scholar] [CrossRef]
- Guthmiller, J.J.; Lan, Y.L.; Fernández-Quintero, M.L.; Han, J.; Utset, H.A.; Bitar, D.J.; Hamel, N.J.; Stovicek, O.; Li, L.; Tepora, M.; et al. Polyreactive broadly neutralizing B cells are selected to provide defense against pandemic threat influenza viruses. Immunity 2020, 53, 1230–1244. [Google Scholar] [CrossRef]
- Prigent, J.; Jarossay, A.; Planchais, C.; Eden, C.; Dufloo, J.; Kök, A.; Lorin, V.; Vratskikh, O.; Couderc, T.; Bruel, T.; et al. Conformational plasticity in broadly neutralizing HIV-1 antibodies triggers polyreactivity. Cell Rep. 2018, 23, 2568–2581. [Google Scholar] [CrossRef] [Green Version]
- Fernández-Quintero, M.L.; Loeffler, J.R.; Kraml, J.; Kahler, U.; Kamenik, A.S.; Liedl, K.R. Characterizing the diversity of the CDR-H3 loop conformational ensembles in relationship to antibody binding properties. Front. Immunol. 2019, 9, 3065. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.J.; Notkins, A.L. Molecular determinants of polyreactive antibody binding: HCDR3 and cyclic peptides. Clin. Exp. Immunol. 2000, 119, 69–76. [Google Scholar] [CrossRef] [PubMed]
- James, L.C.; Roversi, P.; Tawfik, D.S. Antibody multispecificity mediated by conformational diversity. Science 2003, 299, 1362–1367. [Google Scholar] [CrossRef] [Green Version]
- James, L.C.; Tawfik, D.S. The specificity of cross-reactivity: Promiscuous antibody binding involves specific hydrogen bonds rather than nonspecific hydrophobic stickiness. Protein Sci. 2003, 12, 2183–2193. [Google Scholar] [CrossRef] [Green Version]
- Lescar, J.; Pellegrini, M.; Souchon, H.; Tello, D.; Poljak, R.J.; Peterson, N.; Greene, M.; Alzari, P.M. Crystal structure of a cross-reaction complex between Fab F9. 13.7 and guinea fowl lysozyme. J. Biol. Chem. 1995, 270, 18067–18076. [Google Scholar] [CrossRef] [Green Version]
- Tas, J.M.; Mesin, L.; Pasqual, G.; Targ, S.; Jacobsen, J.T.; Mano, Y.M.; Chen, C.S.; Weill, J.C.; Reynaud, C.A.; Browne, E.P.; et al. Visualizing antibody affinity maturation in germinal centers. Science 2016, 351, 1048–1054. [Google Scholar] [CrossRef] [Green Version]
- Kavaler, J.; Caton, A.J.; Staudt, L.M.; Schwartz, D.; Gerhard, W. A set of closely related antibodies dominates the primary antibody response to the antigenic site CB of the A/PR/8/34 influenza virus hemagglutinin. J. Immunol. 1990, 145, 2312–2321. [Google Scholar] [PubMed]
- Angeletti, D.; Gibbs, J.S.; Angel, M.; Kosik, I.; Hickman, H.D.; Frank, G.M.; Das, S.R.; Wheatley, A.K.; Prabhakaran, M.; Leggat, D.J.; et al. Defining B cell immunodominance to viruses. Nat. Immunol. 2017, 18, 456–463. [Google Scholar] [CrossRef] [PubMed]
- Amitai, A.; Sangesland, M.; Barnes, R.M.; Rohrer, D.; Lonberg, N.; Lingwood, D.; Chakraborty, A.K. Defining and manipulating B cell immunodominance hierarchies to elicit broadly neutralizing antibody responses against influenza virus. Cell Syst. 2020, 11, 573–588. [Google Scholar] [CrossRef] [PubMed]
- Altman, M.O.; Angeletti, D.; Yewdell, J.W. Antibody immunodominance: The key to understanding influenza virus antigenic drift. Viral Immunol. 2018, 31, 142–149. [Google Scholar] [CrossRef] [PubMed]
- Steinhauer, D.A.; Domingo, E.; Holland, J.J. Lack of evidence for proofreading mechanisms associated with an RNA virus polymerase. Gene 1992, 122, 281–288. [Google Scholar] [CrossRef]
- Domingo, E.; Holland, J.J. Mutation rates and rapid evolution of RNA viruses. In Evolutionary Biology of Viruses; Morse, S.S., Ed.; Raven Press: New York, NY, USA, 1994; pp. 161–184. [Google Scholar]
- Sanjuán, R.; Nebot, M.R.; Chirico, N.; Mansky, L.M.; Belshaw, R. Viral mutation rates. J. Virol. 2010, 84, 9733–9748. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stray, S.J.; Air, G.M. Apoptosis by influenza viruses correlates with efficiency of viral mRNA synthesis. Virus Res. 2001, 77, 3–17. [Google Scholar] [CrossRef]
- Flipse, J.; Diosa-Toro, M.A.; Hoornweg, T.E.; van de Pol, D.P.I.; Urcuqui-Inchima, S.; Smit, J.M. Antibody-dependent enhancement of dengue virus infection in primary human macrophages; balancing higher fusion against antiviral responses. Sci. Rep. 2016, 6, 29201. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Holmes, E.C. Patterns of intra- and interhost nonsynonymous variation reveal strong purifying selection in dengue virus. J. Virol. 2003, 77, 11296–11298. [Google Scholar] [CrossRef] [Green Version]
- Xue, K.S.; Bloom, J.D. Linking influenza virus evolution within and between human hosts. Virus Evol. 2020, 6, veaa010. [Google Scholar] [CrossRef] [Green Version]
- Purtha, W.E.; Tedder, T.F.; Johnson, S.; Bhattacharya, D.; Diamond, M.S. Memory B cells, but not long-lived plasma cells, possess antigen specificities for viral escape mutants. J. Exp. Med. 2011, 208, 2599–2606. [Google Scholar] [CrossRef]
- Beasley, D.W.; Barrett, A.D. Identification of neutralizing epitopes within structural domain III of the West Nile virus envelope protein. J. Virol. 2002, 76, 13097–13100. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pierson, T.C.; Xu, Q.; Nelson, S.; Oliphant, T.; Nybakken, G.E.; Fremont, D.H.; Diamond, M.S. The stoichiometry of antibody-mediated neutralization and enhancement of West Nile virus infection. Cell Host Microb. 2007, 1, 135–145. [Google Scholar] [CrossRef] [Green Version]
- Slon Campo, J.L.; Mongkolsapaya, J.; Screaton, G.R. The immune response against flaviviruses. Nat. Immunol. 2018, 19, 1189–1198. [Google Scholar] [CrossRef] [PubMed]
- Shinnakasu, R.; Inoue, T.; Kometani, K.; Moriyama, S.; Adachi, Y.; Nakayama, M.; Takahashi, Y.; Fukuyama, H.; Okada, T.; Kurosaki, T. Regulated selection of germinal-center cells into the memory B cell compartment. Nat. Immunol. 2016, 17, 861–869. [Google Scholar] [CrossRef]
- Zhou, D.; Dejnirattisai, W.; Supasa, P.; Liu, C.; Mentzer, A.J.; Ginn, H.M.; Zhao, Y.; Duyvesteyn, H.M.E.; Tuekprakhon, A.; Nutalai, R.; et al. Evidence of escape of SARS-CoV-2 variant B.1.351 from natural and vaccine-induced sera. Cell 2021. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Nair, M.S.; Liu, L.; Iketani, S.; Luo, Y.; Guo, Y.; Wang, M.; Yu, J.; Zhang, B.; Kwong, P.D.; et al. Antibody resistance of SARS-CoV-32 variants B.1.351 and B.1.1.7. Nature 2021. [Google Scholar] [CrossRef] [PubMed]
- Kemp, S.A.; Collier, D.A.; Datir, R.P.; Ferreira, I.A.T.M.; Gayed, S.; Jahun, A.; Hosmillo, M.; Rees-Spear, C.; Mlcochova, P.; Ushiro Lumb, I.; et al. Sars-CoV-2 evolution during treatment of chronic infection. Nature 2021. [Google Scholar] [CrossRef] [PubMed]
- Sabino, E.C.; Buss, L.F.; Carvalho, M.P.S.; Prete, C.A., Jr.; Crispim, M.A.E.; Fraiji, N.A.; Pereira, R.H.M.; Parag, K.V.; da Silva Peixoto, P.; Kraemer, M.U.G.; et al. Resurgence of COVID-19 in Manaus, Brazil, despite high seroprevalence. Lancet 2021, 397, 452–455. [Google Scholar] [CrossRef]
- Gaebler, C.; Wang, Z.; Lorenzi, J.C.C.; Muecksch, F.; Finkin, S.; Tokuyama, M.; Cho, A.; Jankovic, M.; Schaefer-Babajew, D.; Oliveira, T.Y.; et al. Evolutiom of antibody immunity to SARS-CoV-2. Nature 2021. [Google Scholar] [CrossRef] [PubMed]
- Starr, T.N.; Greaney, A.J.; Hilton, S.K.; Ellis, D.; Crawford, K.H.D.; Dingens, A.S.; Navarro, M.J.; Bowen, J.E.; Tortorici, M.A.; Walls, A.C.; et al. Deep mutational scanning of SARS-CoV-2 receptor binding domain reveals constraints on folding and ACE2 binding. Cell 2020, 182, 1295–1310. [Google Scholar] [CrossRef]
- Andreano, E.; Piccini, G.; Licastro, D.; Casalino, L.; Johnson, N.V.; Paciello, I.; Monego, S.D.; Pantano, E.; Manganaro, N.; Manenti, A.; et al. SARS-CoV-2 escape in vitro from a highly neutralizing COVID-19 convalescent plasma. bioRxiv 2020. [Google Scholar] [CrossRef]
- Zahradník, J.; Marciano, S.; Shemesh, M.; Zoler, E.; Chiaravalli, J.; Meyer, B.; Dym, O.; Elad, N.; Schreiber, G. SARS-CoV-2 RBD in vitro evolution follows contagious mutation spread, yet generates an able infection inhibitor. bioRxiv 2021. [Google Scholar] [CrossRef]
- Turner, J.S.; Zhou, J.Q.; Han, J.; Schmitz, A.J.; Rizk, A.A.; Alsoussi, W.B.; Lei, T.L.; Amor, M.; McIntire, K.M.; Meade, P.; et al. Human germinal centres engage memory and naive B cells after influenza vaccination. Nature 2020, 586, 127–132. [Google Scholar] [CrossRef] [PubMed]
- Hahn, Y.S.; Caller, R.; Hunkapiller, T.; Dalrymple, J.M.; Strauss, J.H.; Strauss, E.G. Nucleotide sequence of dengue 2 RNA and comparison of the encoded proteins with those of other flaviviruses. Virology 1988, 162, 167–180. [Google Scholar] [CrossRef]
- Gething, M.J.; Bye, J.; Skehel, J.; Waterfield, M. Cloning and DNA sequence of double-stranded copies of haemagglutinin genes from H2 and H3 strains elucidates antigenic shift and drift in human influenza virus. Nature 1980, 287, 301–306. [Google Scholar] [CrossRef]
- Good, K.L.; Tangye, S.G. Decreased expression of Krüppel-like factors in memory B cells induces the rapid response typical of secondary antibody responses. Proc. Natl. Acad. Sci. USA 2007, 104, 13420–13425. [Google Scholar] [CrossRef] [Green Version]
- Good, K.L.; Avery, D.T.; Tangye, S.G. Resting human memory B cells are intrinsically programmed for enhanced survival and responsiveness to diverse stimuli compared to naive B cells. J. Immunol. 2009, 182, 890–901. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Midgley, C.M.; Bajwa-Joseph, M.; Vasanawathana, S.; Limpitikul, W.; Wills, B.; Flanagan, A.; Waiyaiya, E.; Tran, H.B.; Cowper, A.E.; Chotiyarnwong, P.; et al. An in-depth analysis of original antigenic sin in dengue virus infection. J. Virol. 2011, 85, 410–421. [Google Scholar] [CrossRef] [Green Version]
- Yewdell, J.W.; Santos, J.J.S. Original antigenic sin: How Original? How Sinful? Cold Spring Harb. Perspect. Med. 2020. [Google Scholar] [CrossRef] [Green Version]
- Dejnirattisai, W.; Supasa, P.; Wongwiwat, W.; Rouvinski, A.; Barba-Spaeth, G.; Duangchinda, T.; Sakuntabhai, A.; Cao-Lormeau, V.M.; Malasit, P.; Rey, F.A.; et al. Dengue virus sero-cross-reactivity drives antibody-dependent enhancement of infection with zika virus. Nat. Immunol. 2016, 17, 1102–1108. [Google Scholar] [CrossRef]
- Jardine, J.G.; Kulp, D.W.; Havenar-Daughton, C.; Sarkar, A.; Briney, B.; Sok, D.; Sesterhenn, F.; Ereño-Orbea, J.; Kalyuzhniy, O.; Deresa, I.; et al. HIV-1 broadly neutralizing antibody precursor B cells revealed by germline-targeting immunogen. Science 2016, 351, 1458–1463. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Steichen, J.M.; Lin, Y.C.; Havenar-Daughton, C.; Pecetta, S.; Ozorowski, G.; Willis, J.R.; Toy, L.; Sok, D.; Liguori, A.; Kratochvil, S.; et al. A generalized HIV vaccine design strategy for priming of broadly neutralizing antibody responses. Science 2019, 366, eaax4380. [Google Scholar] [CrossRef] [PubMed]
- Kanekiyo, M.; Joyce, M.G.; Gillespie, R.A.; Gallagher, J.R.; Andrews, S.F.; Yassine, H.M.; Wheatley, A.K.; Fisher, B.E.; Ambrozak, D.R.; Creanga, A.; et al. Mosaic nanoparticle display of diverse influenza virus hemagglutinins elicits broad B cell responses. Nat. Immunol. 2019, 20, 362–372. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
White, H.N. B-Cell Memory Responses to Variant Viral Antigens. Viruses 2021, 13, 565. https://0-doi-org.brum.beds.ac.uk/10.3390/v13040565
White HN. B-Cell Memory Responses to Variant Viral Antigens. Viruses. 2021; 13(4):565. https://0-doi-org.brum.beds.ac.uk/10.3390/v13040565
Chicago/Turabian StyleWhite, Harry N. 2021. "B-Cell Memory Responses to Variant Viral Antigens" Viruses 13, no. 4: 565. https://0-doi-org.brum.beds.ac.uk/10.3390/v13040565