Discriminating between JCPyV and BKPyV in Urinary Virome Data Sets
Abstract
:1. Introduction
2. Materials and Methods
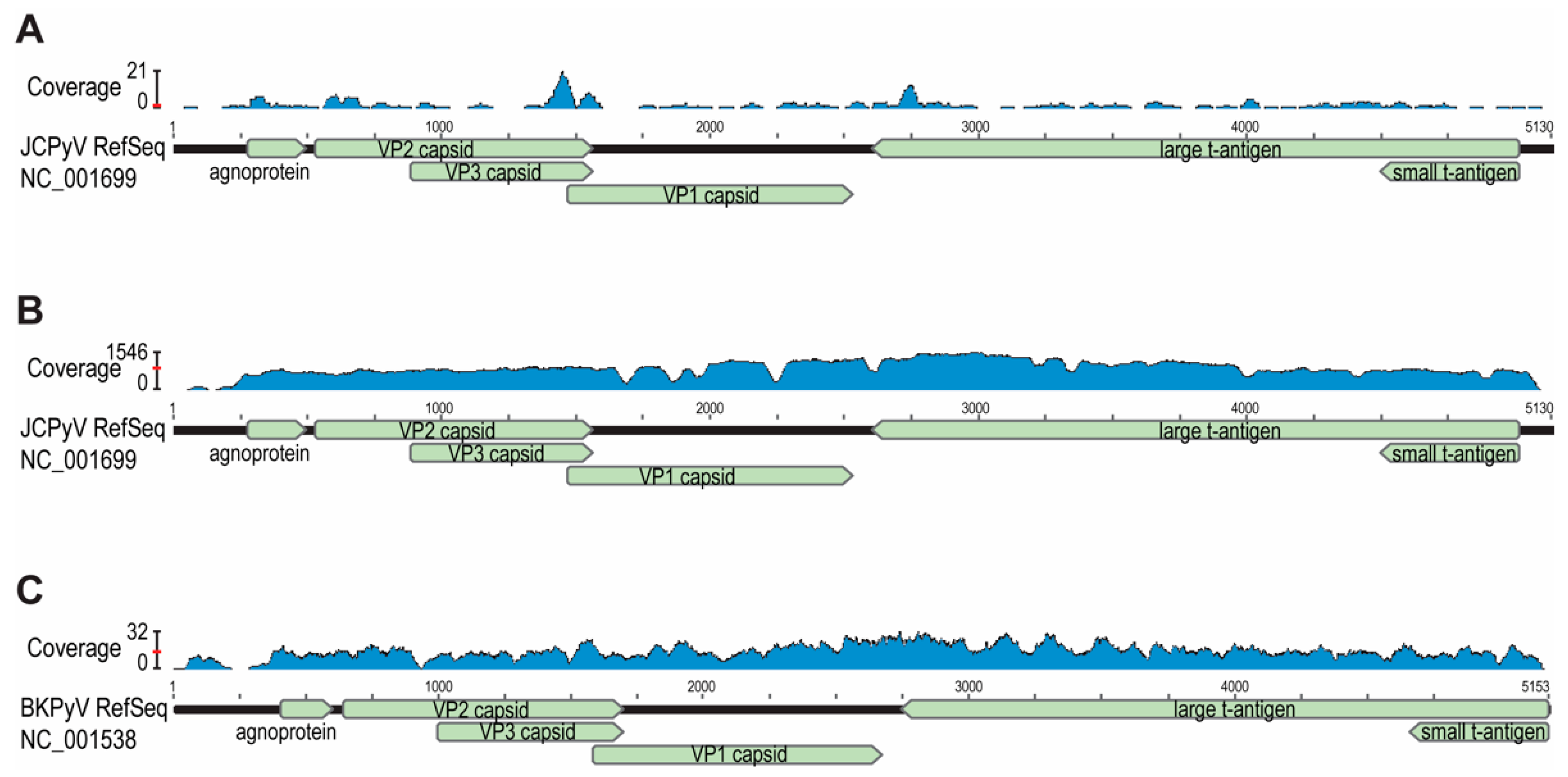
3. Results
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Neugent, M.L.; Hulyalkar, N.V.; Nguyen, V.H.; Zimmern, P.E.; De Nisco, N.J. Advances in Understanding the Human Urinary Microbiome and Its Potential Role in Urinary Tract Infection. mBio 2020, 11. [Google Scholar] [CrossRef]
- Gardner, S.D.; Field, A.M.; Coleman, D.V.; Hulme, B. New Human Papovavirus (B.K.) Isolated from Urine after Renal Transplantation. Lancet 1971, 1, 1253–1257. [Google Scholar] [CrossRef]
- Wylie, K.M.; Mihindukulasuriya, K.A.; Zhou, Y.; Sodergren, E.; Storch, G.A.; Weinstock, G.M. Metagenomic Analysis of Double-Stranded DNA Viruses in Healthy Adults. BMC Biol. 2014, 12, 71. [Google Scholar] [CrossRef]
- Pinto, M.; Dobson, S. BK and JC Virus: A Review. J. Infect. 2014, 68 (Suppl. S1), S2–S8. [Google Scholar] [CrossRef] [PubMed]
- Kitamura, T.; Kunitake, T.; Guo, J.; Tominaga, T.; Kawabe, K.; Yogo, Y. Transmission of the Human Polyomavirus JC Virus Occurs Both within the Family and Outside the Family. J. Clin. Microbiol. 1994, 32, 2359–2363. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Agostini, H.T.; Yanagihara, R.; Davis, V.; Ryschkewitsch, C.F.; Stoner, G.L. Asian Genotypes of JC Virus in Native Americans and in a Pacific Island Population: Markers of Viral Evolution and Human Migration. Proc. Natl. Acad. Sci. USA 1997, 94, 14542–14546. [Google Scholar] [CrossRef] [Green Version]
- Bofill-Mas, S.; Girones, R. Excretion and Transmission of JCV in Human Populations. J. Neurovirol. 2001, 7, 345–349. [Google Scholar] [CrossRef] [Green Version]
- Chang, H.; Wang, M.; Tsai, R.-T.; Lin, H.-S.; Huan, J.-S.; Wang, W.-C.; Chang, D. High Incidence of JC Viruria in JC-Seropositive Older Individuals. J. Neurovirol. 2002, 8, 447–451. [Google Scholar] [CrossRef]
- Agostini, H.T.; Deckhut, A.; Jobes, D.V.; Girones, R.; Schlunck, G.; Prost, M.G.; Frias, C.; Pérez-Trallero, E.; Ryschkewitsch, C.F.; Stoner, G.L. Genotypes of JC Virus in East, Central and Southwest Europe. J. Gen. Virol. 2001, 82, 1221–1331. [Google Scholar] [CrossRef] [Green Version]
- Baksh, F.K.; Finkelstein, S.D.; Swalsky, P.A.; Stoner, G.L.; Ryschkewitsch, C.F.; Randhawa, P. Molecular Genotyping of BK and JC Viruses in Human Polyomavirus-Associated Interstitial Nephritis after Renal Transplantation. Am. J. Kidney Dis. 2001, 38, 354–365. [Google Scholar] [CrossRef]
- Bogdanovic, G.; Brytting, M.; Cinque, P.; Grandien, M.; Fridell, E.; Ljungman, P.; Lönnqvist, B.; Hammarin, A.L. Nested PCR for Detection of BK Virus and JC Virus DNA. Clin. Diagn. Virol. 1994, 2, 211–220. [Google Scholar] [CrossRef]
- Cayres-Vallinoto, I.M.V.; Vallinoto, A.C.R.; Azevedo, V.N.; Machado, L.F.A.; Ishak, M.d.O.G.; Ishak, R. Human JCV Infections as a Bio-Anthropological Marker of the Formation of Brazilian Amazonian Populations. PLoS ONE 2012, 7, e46523. [Google Scholar] [CrossRef] [PubMed]
- De Souza, L.M.; Savassi-Ribas, F.; de Almeida, S.G.S.; da Silva, R.N.N.; Baez, C.F.; Zalis, M.G.; Guimarães, M.A.A.M.; Varella, R.B. A Globally Applicable PCR-Based Detection and Discrimination of BK and JC Polyomaviruses. Rev. Inst. Med. Trop. Sao Paulo 2018, 60, e47. [Google Scholar] [CrossRef] [Green Version]
- Del Valle, L.; Gordon, J.; Assimakopoulou, M.; Enam, S.; Geddes, J.F.; Varakis, J.N.; Katsetos, C.D.; Croul, S.; Khalili, K. Detection of JC Virus DNA Sequences and Expression of the Viral Regulatory Protein T-Antigen in Tumors of the Central Nervous System. Cancer Res. 2001, 61, 4287–4293. [Google Scholar] [PubMed]
- Dumonceaux, T.J.; Mesa, C.; Severini, A. Internally Controlled Triplex Quantitative PCR Assay for Human Polyomaviruses JC and BK. J. Clin. Microbiol. 2008, 46, 2829–2836. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dumoulin, A.; Hirsch, H.H. Reevaluating and Optimizing Polyomavirus BK and JC Real-Time PCR Assays to Detect Rare Sequence Polymorphisms. J. Clin. Microbiol. 2011, 49, 1382–1388. [Google Scholar] [CrossRef] [Green Version]
- Funahashi, Y.; Iwata, S.; Ito, Y.; Kojima, S.; Yoshikawa, T.; Hattori, R.; Gotoh, M.; Nishiyama, Y.; Kimura, H. Multiplex Real-Time PCR Assay for Simultaneous Quantification of BK Polyomavirus, JC Polyomavirus, and Adenovirus DNA. J. Clin. Microbiol. 2010, 48, 825–830. [Google Scholar] [CrossRef] [Green Version]
- Giovannelli, I.; Ciccone, N.; Vaggelli, G.; Malva, N.D.; Torricelli, F.; Rossolini, G.M.; Giannecchini, S. Utility of Droplet Digital PCR for the Quantitative Detection of Polyomavirus JC in Clinical Samples. J. Clin. Virol. 2016, 82, 70–75. [Google Scholar] [CrossRef]
- Haghighi, M.F.; Seyyedi, N.; Farhadi, A.; Zare, F.; Kasraian, L.; Refiei Dehbidi, G.R.; Ranjbaran, R.; Behzad-Behbahani, A. Polyomaviruses BK and JC DNA Infection in Peripheral Blood Cells from Blood Donors. Braz. J. Infect. Dis. 2019, 23, 22–26. [Google Scholar] [CrossRef]
- Hori, R.; Murai, Y.; Tsuneyama, K.; Abdel-Aziz, H.O.; Nomoto, K.; Takahashi, H.; Cheng, C.; Kuchina, T.; Harman, B.V.; Takano, Y. Detection of JC Virus DNA Sequences in Colorectal Cancers in Japan. Virchows Archiv 2005, 447, 723–730. [Google Scholar] [CrossRef]
- Hussain, I.; Tasneem, F.; Umer, M.; Pervaiz, A.; Raza, M.; Arshad, M.I.; Shahzad, N. Specific and Quantitative Detection of Human Polyomaviruses BKPyV and JCPyV in the Healthy Pakistani Population. Virol. J. 2017, 14, 86. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karalic, D.; Lazarevic, I.; Knezevic, A.; Cupic, M.; Jevtovic, D.; Jovanovic, T. Distribution of JC Virus Genotypes among Serbian Patients Infected with HIV and in Healthy Donors. J. Med. Virol. 2014, 86, 411–418. [Google Scholar] [CrossRef]
- Kruzel-Davila, E.; Divers, J.; Russell, G.B.; Kra-Oz, Z.; Cohen, M.S.; Langefeld, C.D.; Ma, L.; Lyles, D.S.; Hicks, P.J.; Skorecki, K.L.; et al. JC Viruria Is Associated with Reduced Risk of Diabetic Kidney Disease. J. Clin. Endocrinol. Metab. 2019, 104, 2286–2294. [Google Scholar] [CrossRef] [PubMed]
- Makvandi, M.; Mombeini, H.; Haghighi, S.B.; Dastoorpoor, M.; Khodadad, N.; Babaahmadi, M.K.; Tabasi, M.; Pirmoradi, R. Molecular Epidemiology of JC Polyomavirus in HIV-Infected Patients and Healthy Individuals from Iran. Braz. J. Microbiol. 2020, 51, 37–43. [Google Scholar] [CrossRef] [PubMed]
- McNees, A.L.; White, Z.S.; Zanwar, P.; Vilchez, R.A.; Butel, J.S. Specific and Quantitative Detection of Human Polyomaviruses BKV, JCV, and SV40 by Real Time PCR. J. Clin. Virol. 2005, 34, 52–62. [Google Scholar] [CrossRef] [PubMed]
- Merlino, C.; Bergallo, M.; Daniele, R.; Ponzi, A.N.; Cavallo, R. BKV-DNA and JCV-DNA Co-Quantification Assay to Evaluate Viral Load in Urine and Serum. Mol. Biotechnol. 2005, 30, 1–8. [Google Scholar] [CrossRef]
- Pal, A.; Sirota, L.; Maudru, T.; Peden, K.; Lewis, A.M. Real-Time, Quantitative PCR Assays for the Detection of Virus-Specific DNA in Samples with Mixed Populations of Polyomaviruses. J. Virol. Methods 2006, 135, 32–42. [Google Scholar] [CrossRef]
- Rafique, A.; Jiang, S.C. Genetic Diversity of Human Polyomavirus JCPyV in Southern California Wastewater. J. Water Health 2008, 6, 533–538. [Google Scholar] [CrossRef] [Green Version]
- Rencic, A.; Gordon, J.; Otte, J.; Curtis, M.; Kovatich, A.; Zoltick, P.; Khalili, K.; Andrews, D. Detection of JC Virus DNA Sequence and Expression of the Viral Oncoprotein, Tumor Antigen, in Brain of Immunocompetent Patient with Oligoastrocytoma. Proc. Natl. Acad. Sci. USA 1996, 93, 7352–7357. [Google Scholar] [CrossRef] [Green Version]
- Ryschkewitsch, C.; Jensen, P.; Hou, J.; Fahle, G.; Fischer, S.; Major, E.O. Comparison of PCR-Southern Hybridization and Quantitative Real-Time PCR for the Detection of JC and BK Viral Nucleotide Sequences in Urine and Cerebrospinal Fluid. J. Virol. Methods 2004, 121, 217–221. [Google Scholar] [CrossRef]
- Sehbani, L.; Kabamba-Mukadi, B.; Vandenbroucke, A.-T.; Bodéus, M.; Goubau, P. Specific and Quantitative Detection of Human Polyomaviruses BKV and JCV by LightCycler Real-Time PCR. J. Clin. Virol. 2006, 36, 159–162. [Google Scholar] [CrossRef]
- Tsai, R.T.; Wang, M.; Ou, W.C.; Lee, Y.L.; Li, S.Y.; Fung, C.Y.; Huang, Y.L.; Tzeng, T.Y.; Chen, Y.; Chang, D. Incidence of JC Viruria Is Higher than That of BK Viruria in Taiwan. J. Med. Virol. 1997, 52, 253–257. [Google Scholar] [CrossRef]
- Santiago-Rodriguez, T.M.; Ly, M.; Bonilla, N.; Pride, D.T. The Human Urine Virome in Association with Urinary Tract Infections. Front. Microbiol. 2015, 6, 14. [Google Scholar] [CrossRef] [Green Version]
- Rani, A.; Ranjan, R.; McGee, H.S.; Metwally, A.; Hajjiri, Z.; Brennan, D.C.; Finn, P.W.; Perkins, D.L. A Diverse Virome in Kidney Transplant Patients Contains Multiple Viral Subtypes with Distinct Polymorphisms. Sci. Rep. 2016, 6. [Google Scholar] [CrossRef]
- Garretto, A.; Thomas-White, K.; Wolfe, A.J.; Putonti, C. Detecting Viral Genomes in the Female Urinary Microbiome. J. Gen. Virol. 2018, 99, 1141–1146. [Google Scholar] [CrossRef]
- Moustafa, A.; Li, W.; Singh, H.; Moncera, K.J.; Torralba, M.G.; Yu, Y.; Manuel, O.; Biggs, W.; Venter, J.C.; Nelson, K.E.; et al. Microbial Metagenome of Urinary Tract Infection. Sci. Rep. 2018, 8. [Google Scholar] [CrossRef]
- Carpenter, M.L.; Tan, S.K.; Watson, T.; Bacher, R.; Nagesh, V.; Watts, A.; Bentley, G.; Weber, J.; Huang, C.; Sahoo, M.K.; et al. Metagenomic Next-Generation Sequencing for Identification and Quantitation of Transplant-Related DNA Viruses. J. Clin. Microbiol. 2019, 57. [Google Scholar] [CrossRef] [Green Version]
- Langmead, B.; Salzberg, S.L. Fast Gapped-Read Alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [Green Version]
- Thomas-White, K.; Forster, S.C.; Kumar, N.; Van Kuiken, M.; Putonti, C.; Stares, M.D.; Hilt, E.E.; Price, T.K.; Wolfe, A.J.; Lawley, T.D. Culturing of Female Bladder Bacteria Reveals an Interconnected Urogenital Microbiota. Nat. Commun. 2018, 9, 1557. [Google Scholar] [CrossRef]
- Wommack, K.E.; Bhavsar, J.; Polson, S.W.; Chen, J.; Dumas, M.; Srinivasiah, S.; Furman, M.; Jamindar, S.; Nasko, D.J. VIROME: A Standard Operating Procedure for Analysis of Viral Metagenome Sequences. Stand. Genom. Sci. 2012, 6, 427–439. [Google Scholar] [CrossRef] [Green Version]
- Huson, D.H.; Weber, N. Microbial Community Analysis Using MEGAN. Methods Enzymol. 2013, 531, 465–485. [Google Scholar] [CrossRef]
- Roux, S.; Tournayre, J.; Mahul, A.; Debroas, D.; Enault, F. Metavir 2: New Tools for Viral Metagenome Comparison and Assembled Virome Analysis. BMC Bioinform. 2014, 15, 76. [Google Scholar] [CrossRef] [Green Version]
- Roux, S.; Enault, F.; Hurwitz, B.L.; Sullivan, M.B. VirSorter: Mining Viral Signal from Microbial Genomic Data. PeerJ 2015, 3, e985. [Google Scholar] [CrossRef] [PubMed]
- Keegan, K.P.; Glass, E.M.; Meyer, F. MG-RAST, a Metagenomics Service for Analysis of Microbial Community Structure and Function. Methods Mol. Biol. 2016, 1399, 207–233. [Google Scholar] [CrossRef]
- Guo, J.; Bolduc, B.; Zayed, A.A.; Varsani, A.; Dominguez-Huerta, G.; Delmont, T.O.; Pratama, A.A.; Gazitúa, M.C.; Vik, D.; Sullivan, M.B.; et al. VirSorter2: A Multi-Classifier, Expert-Guided Approach to Detect Diverse DNA and RNA Viruses. Microbiome 2021, 9, 37. [Google Scholar] [CrossRef] [PubMed]
- Siqueira, J.D.; Curty, G.; Xutao, D.; Hofer, C.B.; Machado, E.S.; Seuánez, H.N.; Soares, M.A.; Delwart, E.; Soares, E.A. Composite Analysis of the Virome and Bacteriome of HIV/HPV Co-Infected Women Reveals Proxies for Immunodeficiency. Viruses 2019, 11, 422. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, H.-T.; Wang, H.; Wu, H.-S.; Zeng, J.; Yang, Y. Comparison of Viromes in Vaginal Secretion from Pregnant Women with and without Vaginitis. Virol. J. 2021, 18, 11. [Google Scholar] [CrossRef]
| Study | Accession No. | # Samples | # Reads Total |
|---|---|---|---|
| Kidney Transplant Virome [34] | PRJEB28510 1 | 27 | 157.5 M 3 |
| Pulmonary Tuberculosis Urinary Microbiome (unpublished) | PRJNA431965 1 | 3 | 9.5 M 4 |
| Microbial Metagenome of UTI [36] | PRJNA385350 1 | 49 | 532.0 M 4 |
| Virome in Healthy and BK Disease of Kidney Transplant (unpublished) | PRJNA587166 1 | 62 | 70.0 M 3 |
| Virome in Association with UTI [33] | cobian9680 2 | 20 | 14.5 M 3 |
| Portable Urinal Microbiome (unpublished) | PRJNA399057 1 | 4 | 228.3 M 4 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mormando, R.; Wolfe, A.J.; Putonti, C. Discriminating between JCPyV and BKPyV in Urinary Virome Data Sets. Viruses 2021, 13, 1041. https://0-doi-org.brum.beds.ac.uk/10.3390/v13061041
Mormando R, Wolfe AJ, Putonti C. Discriminating between JCPyV and BKPyV in Urinary Virome Data Sets. Viruses. 2021; 13(6):1041. https://0-doi-org.brum.beds.ac.uk/10.3390/v13061041
Chicago/Turabian StyleMormando, Rita, Alan J. Wolfe, and Catherine Putonti. 2021. "Discriminating between JCPyV and BKPyV in Urinary Virome Data Sets" Viruses 13, no. 6: 1041. https://0-doi-org.brum.beds.ac.uk/10.3390/v13061041






