From A to m6A: The Emerging Viral Epitranscriptome
Abstract
:1. Introduction
2. N6-Methyladenosine (m6A)
3. m6A Modifications and Viral Replication
4. Epitranscriptomic Regulation of RNA Viruses
4.1. m6A and HIV-1
4.2. m6A and the Flaviviridae Family
4.3. m6A and Chikungunya Virus
4.4. m6A and Coronaviruses
4.5. m6A and Enterovirus 71
4.6. m6A and Influenza A Virus
4.7. m6A and Respiratory Syncytial Virus
4.8. m6A and Vesicular Stomatitis Virus
5. Epitranscriptomic Regulation of DNA Viruses
5.1. m6A and Simian Virus 40
5.2. m6A and Herpesviruses
5.3. m6A and Adenovirus
5.4. m6A and Hepatitis B Virus
6. Epitranscriptomic Regulation of the Immune Response to Viral Infection
7. Conclusions and Future Perspectives
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Davis, F.F.; Allen, F.W. Ribonucleic acids from yeast which contain a fifth nucleotide. J. Biol. Chem. 1957, 227, 907–915. [Google Scholar] [CrossRef]
- Zaccara, S.; Ries, R.J.; Jaffrey, S.R. Reading, writing and erasing mRNA methylation. Nat. Rev. Mol. Cell Biol. 2019, 20, 608–624. [Google Scholar] [CrossRef]
- Desrosiers, R.; Friderici, K.; Rottman, F. Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells. Proc. Natl. Acad. Sci. USA 1974, 71, 3971–3975. [Google Scholar] [CrossRef] [Green Version]
- Perry, R.P.; Kelley, D.E. Existence of methylated messenger RNA in mouse L cells. Cell 1974, 1, 37–42. [Google Scholar] [CrossRef]
- Amos, H.; Korn, M. 5-Methyl cytosine in the RNA of Escherichia coli. Biochim. Biophys. Acta 1958, 29, 444–445. [Google Scholar] [CrossRef]
- Baskin, F.; Dekker, C.A. A Rapid and Specific Assay for Sugar Methylation in Ribonucleic Acid. J. Biol. Chem. 1967, 242, 5447–5449. [Google Scholar] [CrossRef]
- Jia, G.; Fu, Y.; Zhao, X.; Dai, Q.; Zheng, G.; Yang, Y.; Yi, C.; Lindahl, T.; Pan, T.; Yang, Y.-G.; et al. N6-Methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 2011, 7, 885–887. [Google Scholar] [CrossRef]
- Zheng, G.; Dahl John, A.; Niu, Y.; Fedorcsak, P.; Huang, C.-M.; Li Charles, J.; Vågbø Cathrine, B.; Shi, Y.; Wang, W.-L.; Song, S.-H.; et al. ALKBH5 Is a Mammalian RNA Demethylase that Impacts RNA Metabolism and Mouse Fertility. Mol. Cell 2013, 49, 18–29. [Google Scholar] [CrossRef] [Green Version]
- Motorin, Y.; Helm, M. RNA nucleotide methylation. Wires RNA 2011, 2, 611–631. [Google Scholar] [CrossRef] [PubMed]
- Wetzel, C.; Limbach, P.A. Mass spectrometry of modified RNAs: Recent developments. Analyst 2016, 141, 16–23. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, X.; Xiong, X.; Yi, C. Epitranscriptome sequencing technologies: Decoding RNA modifications. Nat. Methods 2017, 14, 23–31. [Google Scholar] [CrossRef]
- Garalde, D.R.; Snell, E.A.; Jachimowicz, D.; Sipos, B.; Lloyd, J.H.; Bruce, M.; Pantic, N.; Admassu, T.; James, P.; Warland, A.; et al. Highly parallel direct RNA sequencing on an array of nanopores. Nat. Methods 2018, 15, 201–206. [Google Scholar] [CrossRef]
- Jonkhout, N.; Tran, J.; Smith, M.A.; Schonrock, N.; Mattick, J.S.; Novoa, E.M. The RNA modification landscape in human disease. RNA 2017, 23, 1754–1769. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roundtree, I.A.; Evans, M.E.; Pan, T.; He, C. Dynamic RNA Modifications in Gene Expression Regulation. Cell 2017, 169, 1187–1200. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Flores, J.V.; Cordero-Espinoza, L.; Oeztuerk-Winder, F.; Andersson-Rolf, A.; Selmi, T.; Blanco, S.; Tailor, J.; Dietmann, S.; Frye, M. Cytosine-5 RNA Methylation Regulates Neural Stem Cell Differentiation and Motility. Stem. Cell Rep. 2017, 8, 112–124. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Livneh, I.; Moshitch-Moshkovitz, S.; Amariglio, N.; Rechavi, G.; Dominissini, D. The m6A epitranscriptome: Transcriptome plasticity in brain development and function. Nat. Rev. Neurosci. 2020, 21, 36–51. [Google Scholar] [CrossRef]
- Batista Pedro, J.; Molinie, B.; Wang, J.; Qu, K.; Zhang, J.; Li, L.; Bouley Donna, M.; Lujan, E.; Haddad, B.; Daneshvar, K.; et al. m6A RNA Modification Controls Cell Fate Transition in Mammalian Embryonic Stem Cells. Cell Stem. Cell 2014, 15, 707–719. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shulman, Z.; Stern-Ginossar, N. The RNA modification N6-methyladenosine as a novel regulator of the immune system. Nat. Immunol. 2020, 21, 501–512. [Google Scholar] [CrossRef] [PubMed]
- Fustin, J.-M.; Doi, M.; Yamaguchi, Y.; Hida, H.; Nishimura, S.; Yoshida, M.; Isagawa, T.; Morioka Masaki, S.; Kakeya, H.; Manabe, I.; et al. RNA-Methylation-Dependent RNA Processing Controls the Speed of the Circadian Clock. Cell 2013, 155, 793–806. [Google Scholar] [CrossRef] [Green Version]
- Bohnsack, K.E.; Höbartner, C.; Bohnsack, M.T. Eukaryotic 5-methylcytosine (m⁵C) RNA Methyltransferases: Mechanisms, Cellular Functions, and Links to Disease. Genes 2019, 10, 102. [Google Scholar] [CrossRef] [Green Version]
- Dai, D.; Wang, H.; Zhu, L.; Jin, H.; Wang, X. N6-methyladenosine links RNA metabolism to cancer progression. Cell Death Dis. 2018, 9, 124. [Google Scholar] [CrossRef] [Green Version]
- Dimitrova, D.G.; Teysset, L.; Carré, C. RNA 2′-O-Methylation (Nm) Modification in Human Diseases. Genes 2019, 10, 117. [Google Scholar] [CrossRef] [Green Version]
- Kadumuri, R.V.; Janga, S.C. Epitranscriptomic Code and Its Alterations in Human Disease. Trends Mol. Med. 2018, 24, 886–903. [Google Scholar] [CrossRef] [PubMed]
- Gonzales-van Horn, S.R.; Sarnow, P. Making the Mark: The Role of Adenosine Modifications in the Life Cycle of RNA Viruses. Cell Host Microbe 2017, 21, 661–669. [Google Scholar] [CrossRef] [Green Version]
- Kennedy, E.M.; Courtney, D.G.; Tsai, K.; Cullen, B.R. Viral Epitranscriptomics. J. Virol. 2017, 91, e02263-16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Netzband, R.; Pager, C.T. Epitranscriptomic marks: Emerging modulators of RNA virus gene expression. Wiley Interdiscip. Rev. RNA 2020, 11, e1576. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tan, B.; Gao, S.-J. RNA epitranscriptomics: Regulation of infection of RNA and DNA viruses by N(6) -methyladenosine (m(6) A). Rev. Med. Virol. 2018, 28, e1983. [Google Scholar] [CrossRef] [Green Version]
- Gokhale, N.S.; McIntyre, A.B.R.; Mattocks, M.D.; Holley, C.L.; Lazear, H.M.; Mason, C.E.; Horner, S.M. Altered m6A Modification of Specific Cellular Transcripts Affects Flaviviridae Infection. Mol. Cell 2020, 77, 542–555.e8. [Google Scholar] [CrossRef] [PubMed]
- Dominissini, D.; Moshitch-Moshkovitz, S.; Schwartz, S.; Salmon-Divon, M.; Ungar, L.; Osenberg, S.; Cesarkas, K.; Jacob-Hirsch, J.; Amariglio, N.; Kupiec, M.; et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 2012, 485, 201–206. [Google Scholar] [CrossRef] [PubMed]
- Meyer, K.D.; Saletore, Y.; Zumbo, P.; Elemento, O.; Mason, C.E.; Jaffrey, S.R. Comprehensive analysis of mRNA methylation reveals enrichment in 3’ UTRs and near stop codons. Cell 2012, 149, 1635–1646. [Google Scholar] [CrossRef] [Green Version]
- Fazi, F.; Fatica, A. Interplay Between N6-Methyladenosine (m6A) and Non-coding RNAs in Cell Development and Cancer. Front. Cell Dev. Biol. 2019, 7, 116. [Google Scholar] [CrossRef] [PubMed]
- Lei, M.; Zheng, G.; Ning, Q.; Zheng, J.; Dong, D. Translation and functional roles of circular RNAs in human cancer. Mol. Cancer 2020, 19, 30. [Google Scholar] [CrossRef]
- Alarcón, C.R.; Goodarzi, H.; Lee, H.; Liu, X.; Tavazoie, S.; Tavazoie, S.F. HNRNPA2B1 Is a Mediator of m6A-Dependent Nuclear RNA Processing Events. Cell 2015, 162, 1299–1308. [Google Scholar] [CrossRef] [Green Version]
- Pendleton, K.E.; Chen, B.; Liu, K.; Hunter, O.V.; Xie, Y.; Tu, B.P.; Conrad, N.K. The U6 snRNA m(6)A Methyltransferase METTL16 Regulates SAM Synthetase Intron Retention. Cell 2017, 169, 824–835.e14. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Yue, Y.; Han, D.; Wang, X.; Fu, Y.; Zhang, L.; Jia, G.; Yu, M.; Lu, Z.; Deng, X.; et al. A METTL3–METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat. Chem. Biol. 2014, 10, 93–95. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Feng, J.; Xue, Y.; Guan, Z.; Zhang, D.; Liu, Z.; Gong, Z.; Wang, Q.; Huang, J.; Tang, C.; et al. Structural basis of N6-adenosine methylation by the METTL3–METTL14 complex. Nature 2016, 534, 575–578. [Google Scholar] [CrossRef] [PubMed]
- Geula, S.; Moshitch-Moshkovitz, S.; Dominissini, D.; Mansour, A.A.; Kol, N.; Salmon-Divon, M.; Hershkovitz, V.; Peer, E.; Mor, N.; Manor, Y.S.; et al. m6A mRNA methylation facilitates resolution of naïve pluripotency toward differentiation. Science 2015, 347, 1002. [Google Scholar] [CrossRef] [PubMed]
- Ping, X.-L.; Sun, B.-F.; Wang, L.; Xiao, W.; Yang, X.; Wang, W.-J.; Adhikari, S.; Shi, Y.; Lv, Y.; Chen, Y.-S.; et al. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. 2014, 24, 177–189. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Linder, B.; Grozhik, A.V.; Olarerin-George, A.O.; Meydan, C.; Mason, C.E.; Jaffrey, S.R. Single-nucleotide-resolution mapping of m6A and m6Am throughout the transcriptome. Nat. Methods 2015, 12, 767–772. [Google Scholar] [CrossRef]
- Mauer, J.; Jaffrey, S.R. FTO, m6Am, and the hypothesis of reversible epitranscriptomic mRNA modifications. FEBS Lett. 2018, 592, 2012–2022. [Google Scholar] [CrossRef] [Green Version]
- Mauer, J.; Luo, X.; Blanjoie, A.; Jiao, X.; Grozhik, A.V.; Patil, D.P.; Linder, B.; Pickering, B.F.; Vasseur, J.J.; Chen, Q.; et al. Reversible methylation of m6Am in the 5’ cap controls mRNA stability. Nature 2017, 541, 371–375. [Google Scholar] [CrossRef] [Green Version]
- Liao, S.; Sun, H.; Xu, C. YTH Domain: A Family of N6-methyladenosine (m6A) Readers. Genom. Proteom. Bioinform. 2018, 16, 99–107. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Liu, K.; Ahmed, H.; Loppnau, P.; Schapira, M.; Min, J. Structural Basis for the Discriminative Recognition of N6-Methyladenosine RNA by the Human YT521-B Homology Domain Family of Proteins. J. Biol. Chem. 2015, 290, 24902–24913. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, C.; Wang, X.; Liu, K.; Roundtree, I.A.; Tempel, W.; Li, Y.; Lu, Z.; He, C.; Min, J. Structural basis for selective binding of m6A RNA by the YTHDC1 YTH domain. Nat. Chem. Biol. 2014, 10, 927–929. [Google Scholar] [CrossRef]
- Dai, X.; Wang, T.; Gonzalez, G.; Wang, Y. Identification of YTH Domain-Containing Proteins as the Readers for N1-Methyladenosine in RNA. Anal. Chem. 2018, 90, 6380–6384. [Google Scholar] [CrossRef] [PubMed]
- Du, H.; Zhao, Y.; He, J.; Zhang, Y.; Xi, H.; Liu, M.; Ma, J.; Wu, L. YTHDF2 destabilizes m6A-containing RNA through direct recruitment of the CCR4–NOT deadenylase complex. Nat. Commun. 2016, 7, 12626. [Google Scholar] [CrossRef]
- Shi, H.; Wang, X.; Lu, Z.; Zhao, B.S.; Ma, H.; Hsu, P.J.; Liu, C.; He, C. YTHDF3 facilitates translation and decay of N6-methyladenosine-modified RNA. Cell Res. 2017, 27, 315–328. [Google Scholar] [CrossRef] [Green Version]
- Zaccara, S.; Jaffrey, S.R. A Unified Model for the Function of YTHDF Proteins in Regulating m6A-Modified mRNA. Cell 2020, 181, 1582–1595.e8. [Google Scholar] [CrossRef]
- Wang, X.; Zhao, B.S.; Roundtree, I.A.; Lu, Z.; Han, D.; Ma, H.; Weng, X.; Chen, K.; Shi, H.; He, C. N6-methyladenosine Modulates Messenger RNA Translation Efficiency. Cell 2015, 161, 1388–1399. [Google Scholar] [CrossRef] [Green Version]
- Meyer Kate, D.; Patil Deepak, P.; Zhou, J.; Zinoviev, A.; Skabkin Maxim, A.; Elemento, O.; Pestova Tatyana, V.; Qian, S.-B.; Jaffrey Samie, R. 5′ UTR m6A Promotes Cap-Independent Translation. Cell 2015, 163, 999–1010. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Lu, Z.; Gomez, A.; Hon, G.C.; Yue, Y.; Han, D.; Fu, Y.; Parisien, M.; Dai, Q.; Jia, G.; et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 2014, 505, 117–120. [Google Scholar] [CrossRef] [PubMed]
- Li, A.; Chen, Y.-S.; Ping, X.-L.; Yang, X.; Xiao, W.; Yang, Y.; Sun, H.-Y.; Zhu, Q.; Baidya, P.; Wang, X.; et al. Cytoplasmic m6A reader YTHDF3 promotes mRNA translation. Cell Res. 2017, 27, 444–447. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiao, W.; Adhikari, S.; Dahal, U.; Chen, Y.-S.; Hao, Y.-J.; Sun, B.-F.; Sun, H.-Y.; Li, A.; Ping, X.-L.; Lai, W.-Y.; et al. Nuclear m6A Reader YTHDC1 Regulates mRNA Splicing. Mol. Cell 2016, 61, 507–519. [Google Scholar] [CrossRef] [Green Version]
- Roundtree, I.A.; Luo, G.Z.; Zhang, Z.; Wang, X.; Zhou, T.; Cui, Y.; Sha, J.; Huang, X.; Guerrero, L.; Xie, P.; et al. YTHDC1 mediates nuclear export of N(6)-methyladenosine methylated mRNAs. Elife 2017, 6, e31311. [Google Scholar] [CrossRef] [PubMed]
- Mao, Y.; Dong, L.; Liu, X.-M.; Guo, J.; Ma, H.; Shen, B.; Qian, S.-B. m6A in mRNA coding regions promotes translation via the RNA helicase-containing YTHDC2. Nat. Commun. 2019, 10, 5332. [Google Scholar] [CrossRef]
- Bailey, A.S.; Batista, P.J.; Gold, R.S.; Chen, Y.G.; de Rooij, D.G.; Chang, H.Y.; Fuller, M.T. The conserved RNA helicase YTHDC2 regulates the transition from proliferation to differentiation in the germline. Elife 2017, 6, e26116. [Google Scholar] [CrossRef]
- Hsu, P.J.; Zhu, Y.; Ma, H.; Guo, Y.; Shi, X.; Liu, Y.; Qi, M.; Lu, Z.; Shi, H.; Wang, J.; et al. Ythdc2 is an N6-methyladenosine binding protein that regulates mammalian spermatogenesis. Cell Res. 2017, 27, 1115–1127. [Google Scholar] [CrossRef]
- Huang, H.; Weng, H.; Sun, W.; Qin, X.; Shi, H.; Wu, H.; Zhao, B.S.; Mesquita, A.; Liu, C.; Yuan, C.L.; et al. Recognition of RNA N(6)-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat. Cell Biol. 2018, 20, 285–295. [Google Scholar] [CrossRef]
- Baquero-Perez, B.; Antanaviciute, A.; Yonchev, I.D.; Carr, I.M.; Wilson, S.A.; Whitehouse, A. The Tudor SND1 protein is an m6A RNA reader essential for replication of Kaposi’s sarcoma-associated herpesvirus. Elife 2019, 8, e47261. [Google Scholar] [CrossRef]
- Edupuganti, R.R.; Geiger, S.; Lindeboom, R.G.H.; Shi, H.; Hsu, P.J.; Lu, Z.; Wang, S.-Y.; Baltissen, M.P.A.; Jansen, P.W.T.C.; Rossa, M.; et al. N6-methyladenosine (m6A) recruits and repels proteins to regulate mRNA homeostasis. Nat. Struct. Mol. Biol. 2017, 24, 870–878. [Google Scholar] [CrossRef]
- Sun, L.; Fazal, F.M.; Li, P.; Broughton, J.P.; Lee, B.; Tang, L.; Huang, W.; Kool, E.T.; Chang, H.Y.; Zhang, Q.C. RNA structure maps across mammalian cellular compartments. Nat. Struct. Mol. Biol. 2019, 26, 322–330. [Google Scholar] [CrossRef]
- Liu, N.; Dai, Q.; Zheng, G.; He, C.; Parisien, M.; Pan, T. N6-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature 2015, 518, 560–564. [Google Scholar] [CrossRef] [Green Version]
- Liu, N.; Zhou, K.I.; Parisien, M.; Dai, Q.; Diatchenko, L.; Pan, T. N6-methyladenosine alters RNA structure to regulate binding of a low-complexity protein. Nucleic Acids Res. 2017, 45, 6051–6063. [Google Scholar] [CrossRef] [Green Version]
- Hsu, P.J.; Shi, H.; Zhu, A.C.; Lu, Z.; Miller, N.; Edens, B.M.; Ma, Y.C.; He, C. The RNA-binding protein FMRP facilitates the nuclear export of N6-methyladenosine–containing mRNAs. J. Biol. Chem. 2019, 294, 19889–19895. [Google Scholar] [CrossRef] [PubMed]
- Luo, S.; Tong, L. Molecular basis for the recognition of methylated adenines in RNA by the eukaryotic YTH domain. Proc. Natl. Acad. Sci. USA 2014, 111, 13834. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, B.; Su, S.; Patil, D.P.; Liu, H.; Gan, J.; Jaffrey, S.R.; Ma, J. Molecular basis for the specific and multivariant recognitions of RNA substrates by human hnRNP A2/B1. Nat. Commun. 2018, 9, 420. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gokhale, N.S.; McIntyre, A.B.R.; McFadden, M.J.; Roder, A.E.; Kennedy, E.M.; Gandara, J.A.; Hopcraft, S.E.; Quicke, K.M.; Vazquez, C.; Willer, J.; et al. N6-Methyladenosine in Flaviviridae Viral RNA Genomes Regulates Infection. Cell Host Microbe 2016, 20, 654–665. [Google Scholar] [CrossRef] [Green Version]
- Lichinchi, G.; Zhao, B.S.; Wu, Y.; Lu, Z.; Qin, Y.; He, C.; Rana, T.M. Dynamics of Human and Viral RNA Methylation during Zika Virus Infection. Cell Host Microbe 2016, 20, 666–673. [Google Scholar] [CrossRef] [Green Version]
- Wei, J.; Liu, F.; Lu, Z.; Fei, Q.; Ai, Y.; He, P.C.; Shi, H.; Cui, X.; Su, R.; Klungland, A.; et al. Differential m6A, m6Am, and m1A Demethylation Mediated by FTO in the Cell Nucleus and Cytoplasm. Mol. Cell 2018, 71, 973–985.e975. [Google Scholar] [CrossRef] [Green Version]
- Xue, M.; Zhao, B.S.; Zhang, Z.; Lu, M.; Harder, O.; Chen, P.; Lu, Z.; Li, A.; Ma, Y.; Xu, Y.; et al. Viral N6-methyladenosine upregulates replication and pathogenesis of human respiratory syncytial virus. Nat. Commun. 2019, 10, 4595. [Google Scholar] [CrossRef]
- Gulati, P.; Avezov, E.; Ma, M.; Antrobus, R.; Lehner, P.; O’Rahilly, S.; Yeo, G.S.H. Fat mass and obesity-related (FTO) shuttles between the nucleus and cytoplasm. Biosci. Rep. 2014, 34, e00144. [Google Scholar] [CrossRef] [PubMed]
- Lesbirel, S.; Viphakone, N.; Parker, M.; Parker, J.; Heath, C.; Sudbery, I.; Wilson, S.A. The m6A-methylase complex recruits TREX and regulates mRNA export. Sci. Rep. 2018, 8, 13827. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, J.; Wan, J.; Gao, X.; Zhang, X.; Jaffrey, S.R.; Qian, S.-B. Dynamic m6A mRNA methylation directs translational control of heat shock response. Nature 2015, 526, 591–594. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anders, M.; Chelysheva, I.; Goebel, I.; Trenkner, T.; Zhou, J.; Mao, Y.; Ignatova, Z. Dynamic m6A methylation facilitates mRNA triaging to stress granules. Life Sci. Alliance 2018, 1, e201800113. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Li, Y.; Toth, J.I.; Petroski, M.D.; Zhang, Z.; Zhao, J.C. N6-methyladenosine modification destabilizes developmental regulators in embryonic stem cells. Nat. Cell Biol. 2014, 16, 191–198. [Google Scholar] [CrossRef]
- Zhao, X.; Yang, Y.; Sun, B.-F.; Shi, Y.; Yang, X.; Xiao, W.; Hao, Y.-J.; Ping, X.-L.; Chen, Y.-S.; Wang, W.-J.; et al. FTO-dependent demethylation of N6-methyladenosine regulates mRNA splicing and is required for adipogenesis. Cell Res. 2014, 24, 1403–1419. [Google Scholar] [CrossRef]
- He, L.; Li, H.; Wu, A.; Peng, Y.; Shu, G.; Yin, G. Functions of N6-methyladenosine and its role in cancer. Mol. Cancer 2019, 18, 176. [Google Scholar] [CrossRef] [Green Version]
- Paramasivam, A.; Priyadharsini, J.V.; Raghunandhakumar, S. Implications of m6A modification in autoimmune disorders. Cell. Mol. Immunol. 2020, 17, 550–551. [Google Scholar] [CrossRef]
- Noack, F.; Calegari, F. Epitranscriptomics: A New Regulatory Mechanism of Brain Development and Function. Front. Neurosci. 2018, 12, 85. [Google Scholar] [CrossRef]
- Manners, O.; Baquero-Perez, B.; Whitehouse, A. m6A: Widespread regulatory control in virus replication. Biochim. Et Biophys. Acta (Bba) Gene Regul. Mech. 2019, 1862, 370–381. [Google Scholar] [CrossRef]
- McIntyre, A.B.R.; Gokhale, N.S.; Cerchietti, L.; Jaffrey, S.R.; Horner, S.M.; Mason, C.E. Limits in the detection of m6A changes using MeRIP/m6A-seq. Sci. Rep. 2020, 10, 6590. [Google Scholar] [CrossRef] [Green Version]
- Chen, K.; Lu, Z.; Wang, X.; Fu, Y.; Luo, G.-Z.; Liu, N.; Han, D.; Dominissini, D.; Dai, Q.; Pan, T.; et al. High-resolution N6-methyladenosine m6A map using photo-crosslinking-assisted m6A sequencing. Angew. Chem. Int. Ed. Engl. 2015, 54, 1587–1590. [Google Scholar] [CrossRef] [Green Version]
- Molinie, B.; Wang, J.; Lim, K.S.; Hillebrand, R.; Lu, Z.-X.; Van Wittenberghe, N.; Howard, B.D.; Daneshvar, K.; Mullen, A.C.; Dedon, P.; et al. m6A-LAIC-seq reveals the census and complexity of the m6A epitranscriptome. Nat. Methods 2016, 13, 692–698. [Google Scholar] [CrossRef] [PubMed]
- Mikutis, S.; Gu, M.; Sendinc, E.; Hazemi, M.E.; Kiely-Collins, H.; Aspris, D.; Vassiliou, G.S.; Shi, Y.; Tzelepis, K.; Bernardes, G.J.L. meCLICK-Seq, a Substrate-Hijacking and RNA Degradation Strategy for the Study of RNA Methylation. ACS Cent Sci. 2020, 6, 2196–2208. [Google Scholar] [CrossRef] [PubMed]
- Meyer, K.D. DART-seq: An antibody-free method for global m6A detection. Nat. Methods 2019, 16, 1275–1280. [Google Scholar] [CrossRef]
- Garcia-Campos, M.A.; Edelheit, S.; Toth, U.; Safra, M.; Shachar, R.; Viukov, S.; Winkler, R.; Nir, R.; Lasman, L.; Brandis, A.; et al. Deciphering the “m6A Code” via Antibody-Independent Quantitative Profiling. Cell 2019, 178, 731–747.e716. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Xiao, Y.; Dong, S.; Yu, Q.; Jia, G. Antibody-free enzyme-assisted chemical approach for detection of N6-methyladenosine. Nat. Chem. Biol. 2020, 16, 896–903. [Google Scholar] [CrossRef] [PubMed]
- Shu, X.; Cao, J.; Cheng, M.; Xiang, S.; Gao, M.; Li, T.; Ying, X.; Wang, F.; Yue, Y.; Lu, Z.; et al. A metabolic labeling method detects m6A transcriptome-wide at single base resolution. Nat. Chem. Biol. 2020, 16, 887–895. [Google Scholar] [CrossRef] [PubMed]
- Anreiter, I.; Mir, Q.; Simpson, J.T.; Janga, S.C.; Soller, M. New Twists in Detecting mRNA Modification Dynamics. Trends Biotechnol. 2021, 39, 72–89. [Google Scholar] [CrossRef]
- Xu, L.; Seki, M. Recent advances in the detection of base modifications using the Nanopore sequencer. J. Hum. Genet. 2020, 65, 25–33. [Google Scholar] [CrossRef] [Green Version]
- Kim, D.; Lee, J.-Y.; Yang, J.-S.; Kim, J.W.; Kim, V.N.; Chang, H. The Architecture of SARS-CoV-2 Transcriptome. Cell 2020, 181, 914–921.e910. [Google Scholar] [CrossRef]
- Viehweger, A.; Krautwurst, S.; Lamkiewicz, K.; Madhugiri, R.; Ziebuhr, J.; Hölzer, M.; Marz, M. Direct RNA nanopore sequencing of full-length coronavirus genomes provides novel insights into structural variants and enables modification analysis. Genome Res. 2019, 29, 1545–1554. [Google Scholar] [CrossRef] [Green Version]
- Price, A.M.; Hayer, K.E.; McIntyre, A.B.; Gokhale, N.S.; Abebe, J.S.; Della Fera, A.N.; Weitzman, M.D. Direct RNA sequencing reveals m6A modifications on adenovirus RNA are necessary for efficient splicing. Nat. Commun. 2020, 11, 6016. [Google Scholar] [CrossRef]
- Lavi, S.; Shatkin, A.J. Methylated simian virus 40-specific RNA from nuclei and cytoplasm of infected BSC-1 cells. Proc. Natl. Acad. Sci. USA 1975, 72, 2012–2016. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hashimoto, S.I.; Green, M. Multiple methylated cap sequences in adenovirus type 2 early mRNA. J. Virol. 1976, 20, 425–435. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krug, R.M.; Morgan, M.A.; Shatkin, A.J. Influenza viral mRNA contains internal N6-methyladenosine and 5’-terminal 7-methylguanosine in cap structures. J. Virol. 1976, 20, 45–53. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Narayan, P.; Ayers, D.F.; Rottman, F.M.; Maroney, P.A.; Nilsen, T.W. Unequal distribution of N6-methyladenosine in influenza virus mRNAs. Mol. Cell Biol. 1987, 7, 1572–1575. [Google Scholar] [CrossRef] [Green Version]
- Moss, B.; Gershowitz, A.; Stringer, J.R.; Holland, L.E.; Wagner, E.K. 5′-Terminal and internal methylated nucleosides in herpes simplex virus type 1 mRNA. J. Virol. 1977, 23, 234–239. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stoltzfus, C.M.; Dimock, K. Evidence of methylation of B77 avian sarcoma virus genome RNA subunits. J. Virol. 1976, 18, 586–595. [Google Scholar] [CrossRef] [Green Version]
- Beemon, K.; Keith, J. Localization of N6-methyladenosine in the Rous sarcoma virus genome. J. Mol. Biol. 1977, 113, 165–179. [Google Scholar] [CrossRef]
- Kane, S.E.; Beemon, K. Precise localization of m6A in Rous sarcoma virus RNA reveals clustering of methylation sites: Implications for RNA processing. Mol. Cell Biol. 1985, 5, 2298–2306. [Google Scholar] [CrossRef] [Green Version]
- Hesser, C.R.; Karijolich, J.; Dominissini, D.; He, C.; Glaunsinger, B.A. N6-methyladenosine modification and the YTHDF2 reader protein play cell type specific roles in lytic viral gene expression during Kaposi’s sarcoma-associated herpesvirus infection. PLoS Pathog. 2018, 14, e1006995. [Google Scholar] [CrossRef] [Green Version]
- Lichinchi, G.; Gao, S.; Saletore, Y.; Gonzalez, G.M.; Bansal, V.; Wang, Y.; Mason, C.E.; Rana, T.M. Dynamics of the human and viral m6A RNA methylomes during HIV-1 infection of T cells. Nat. Microbiol. 2016, 1, 16011. [Google Scholar] [CrossRef]
- Kennedy, E.M.; Bogerd, H.P.; Kornepati, A.V.R.; Kang, D.; Ghoshal, D.; Marshall, J.B.; Poling, B.C.; Tsai, K.; Gokhale, N.S.; Horner, S.M.; et al. Posttranscriptional m6A Editing of HIV-1 mRNAs Enhances Viral Gene Expression. Cell Host Microbe 2016, 19, 675–685. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, W.; Tirumuru, N.; St Gelais, C.; Koneru, P.C.; Liu, C.; Kvaratskhelia, M.; He, C.; Wu, L. N6-Methyladenosine–binding proteins suppress HIV-1 infectivity and viral production. J. Biol. Chem. 2018, 293, 12992–13005. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tirumuru, N.; Zhao, B.S.; Lu, W.; Lu, Z.; He, C.; Wu, L. N6-methyladenosine of HIV-1 RNA regulates viral infection and HIV-1 Gag protein expression. Elife 2016, 5, e15528. [Google Scholar] [CrossRef]
- Jurczyszak, D.; Zhang, W.; Terry, S.N.; Kehrer, T.; Bermúdez González, M.C.; McGregor, E.; Mulder, L.C.F.; Eckwahl, M.J.; Pan, T.; Simon, V. HIV protease cleaves the antiviral m6A reader protein YTHDF3 in the viral particle. PLoS Pathog. 2020, 16, e1008305. [Google Scholar] [CrossRef] [Green Version]
- Chen, J.; Jin, L.; Wang, Z.; Wang, L.; Chen, Q.; Cui, Y.; Liu, G. N6-methyladenosine regulates PEDV replication and host gene expression. Virology 2020, 548, 59–72. [Google Scholar] [CrossRef] [PubMed]
- Courtney, D.G.; Kennedy, E.M.; Dumm, R.E.; Bogerd, H.P.; Tsai, K.; Heaton, N.S.; Cullen, B.R. Epitranscriptomic Enhancement of Influenza A Virus Gene Expression and Replication. Cell Host Microbe 2017, 22, 377–386.e375. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, J.; Xu, Y.P.; Li, K.; Ye, Q.; Zhou, H.Y.; Sun, H.; Li, X.; Qin, C.F. The m6A methylome of SARS-CoV-2 in host cells. Cell Res. 2021, 31, 404–414. [Google Scholar] [CrossRef] [PubMed]
- Kim, B.; Arcos, S.; Rothamel, K.; Jian, J.; Rose, K.L.; McDonald, W.H.; Bian, Y.; Reasoner, S.; Barrows, N.J.; Bradrick, S.; et al. Discovery of Widespread Host Protein Interactions with the Pre-replicated Genome of CHIKV Using VIR-CLASP. Mol. Cell 2020, 78, 624–640.e627. [Google Scholar] [CrossRef] [PubMed]
- Hao, H.; Hao, S.; Chen, H.; Chen, Z.; Zhang, Y.; Wang, J.; Wang, H.; Zhang, B.; Qiu, J.; Deng, F.; et al. N6-methyladenosine modification and METTL3 modulate enterovirus 71 replication. Nucleic. Acids Res. 2019, 47, 362–374. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, M.; Zhang, Z.; Xue, M.; Zhao, B.S.; Harder, O.; Li, A.; Liang, X.; Gao, T.Z.; Xu, Y.; Zhou, J.; et al. N6-methyladenosine modification enables viral RNA to escape recognition by RNA sensor RIG-I. Nat. Microbiol. 2020, 5, 584–598. [Google Scholar] [CrossRef] [PubMed]
- Qiu, W.; Zhang, Q.; Zhang, R.; Lu, Y.; Wang, X.; Tian, H.; Yang, P. N6-methyladenosine RNA modification suppresses antiviral innate sensing pathways via reshaping double-stranded RNA. Nat. Commun. 2021, 12, 1582. [Google Scholar] [CrossRef]
- Tsai, K.; Courtney, D.G.; Cullen, B.R. Addition of m6A to SV40 late mRNAs enhances viral structural gene expression and replication. PLoS Pathog. 2018, 14, e1006919. [Google Scholar] [CrossRef] [PubMed]
- Ye, F.; Chen, E.R.; Nilsen, T.W. Kaposi’s Sarcoma-Associated Herpesvirus Utilizes and Manipulates RNA N6-Adenosine Methylation To Promote Lytic Replication. J. Virol. 2017, 91, e00466-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tan, B.; Liu, H.; Zhang, S.; da Silva, S.R.; Zhang, L.; Meng, J.; Cui, X.; Yuan, H.; Sorel, O.; Zhang, S.-W.; et al. Viral and cellular N6-methyladenosine and N6,2′-O-dimethyladenosine epitranscriptomes in the KSHV life cycle. Nat. Microbiol. 2018, 3, 108–120. [Google Scholar] [CrossRef]
- Lang, F.; Singh, R.K.; Pei, Y.; Zhang, S.; Sun, K.; Robertson, E.S. EBV epitranscriptome reprogramming by METTL14 is critical for viral-associated tumorigenesis. PLoS Pathog. 2019, 15, e1007796. [Google Scholar] [CrossRef]
- Xia, T.L.; Li, X.; Wang, X.; Zhu, Y.J.; Zhang, H.; Cheng, W.; Zeng, M.S. N(6)-methyladenosine-binding protein YTHDF1 suppresses EBV replication and promotes EBV RNA decay. EMBO Rep. 2021, 22, e50128. [Google Scholar] [CrossRef]
- Imam, H.; Khan, M.; Gokhale, N.S.; McIntyre, A.B.R.; Kim, G.-W.; Jang, J.Y.; Kim, S.-J.; Mason, C.E.; Horner, S.M.; Siddiqui, A. N6-methyladenosine modification of hepatitis B virus RNA differentially regulates the viral life cycle. Proc. Natl. Acad. Sci. USA 2018, 115, 8829–8834. [Google Scholar] [CrossRef] [Green Version]
- Winkler, R.; Gillis, E.; Lasman, L.; Safra, M.; Geula, S.; Soyris, C.; Nachshon, A.; Tai-Schmiedel, J.; Friedman, N.; Le-Trilling, V.T.K.; et al. m6A modification controls the innate immune response to infection by targeting type I interferons. Nat. Immunol. 2019, 20, 173–182. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; You, Y.; Lu, Z.; Yang, J.; Li, P.; Liu, L.; Xu, H.; Niu, Y.; Cao, X. N6-methyladenosine RNA modification–mediated cellular metabolism rewiring inhibits viral replication. Science 2019, 365, 1171–1176. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wang, X.; Zhang, X.; Wang, J.; Ma, Y.; Zhang, L.; Cao, X. RNA-binding protein YTHDF3 suppresses interferon-dependent antiviral responses by promoting FOXO3 translation. Proc. Natl. Acad. Sci. USA 2019, 116, 976–981. [Google Scholar] [CrossRef] [Green Version]
- Rubio, R.M.; Depledge, D.P.; Bianco, C.; Thompson, L.; Mohr, I. RNA m6A modification enzymes shape innate responses to DNA by regulating interferon β. Genes Dev. 2018, 32, 1472–1484. [Google Scholar] [CrossRef] [Green Version]
- Tirumuru, N.; Wu, L. HIV-1 envelope proteins up-regulate N (6)-methyladenosine levels of cellular RNA independently of viral replication. J. Biol. Chem. 2019, 294, 3249–3260. [Google Scholar] [CrossRef] [Green Version]
- Chu, C.-C.; Liu, B.; Plangger, R.; Kreutz, C.; Al-Hashimi, H.M. m6A minimally impacts the structure, dynamics, and Rev ARM binding properties of HIV-1 RRE stem IIB. PLoS ONE 2019, 14, e0224850. [Google Scholar] [CrossRef] [Green Version]
- Kim, G.-W.; Imam, H.; Khan, M.; Siddiqui, A. N6-Methyladenosine modification of hepatitis B and C viral RNAs attenuates host innate immunity via RIG-I signaling. J. Biol. Chem. 2020, 295, 13123–13133. [Google Scholar] [CrossRef]
- Antanaviciute, A.; Baquero-Perez, B.; Watson, C.M.; Harrison, S.M.; Lascelles, C.; Crinnion, L.; Markham, A.F.; Bonthron, D.T.; Whitehouse, A.; Carr, I.M. m6aViewer: Software for the detection, analysis, and visualization of N6-methyladenosine peaks from m6A-seq/ME-RIP sequencing data. RNA 2017, 23, 1493–1501. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.; Nott, T.J.; Jin, J.; Pawson, T. Deciphering arginine methylation: Tudor tells the tale. Nat. Rev. Mol. Cell Biol. 2011, 12, 629–642. [Google Scholar] [CrossRef] [PubMed]
- Meng, Y.; Li, S.; Gu, D.; Xu, K.; Du, M.; Zhu, L.; Chu, H.; Zhang, Z.; Wu, Y.; Fu, Z.; et al. Genetic variants in m6A modification genes are associated with colorectal cancer risk. Carcinogenesis 2019, 41, 8–17. [Google Scholar] [CrossRef]
- Ivashkiv, L.B.; Donlin, L.T. Regulation of type I interferon responses. Nat. Rev. Immunol. 2014, 14, 36–49. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, H.; Wang, X.; Cai, J.; Dai, Q.; Natchiar, S.K.; Lv, R.; Chen, K.; Lu, Z.; Chen, H.; Shi, Y.G.; et al. N6-Methyladenosine methyltransferase ZCCHC4 mediates ribosomal RNA methylation. Nat. Chem. Biol. 2019, 15, 88–94. [Google Scholar] [CrossRef]
- van Tran, N.; Ernst, F.G.M.; Hawley, B.R.; Zorbas, C.; Ulryck, N.; Hackert, P.; Bohnsack, K.E.; Bohnsack, M.T.; Jaffrey, S.R.; Graille, M.; et al. The human 18S rRNA m6A methyltransferase METTL5 is stabilized by TRMT112. Nucleic Acids Res. 2019, 47, 7719–7733. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Z.; Theler, D.; Kaminska, K.H.; Hiller, M.; de la Grange, P.; Pudimat, R.; Rafalska, I.; Heinrich, B.; Bujnicki, J.M.; Allain, F.H.T.; et al. The YTH domain is a novel RNA binding domain. J. Biol. Chem. 2010, 285, 14701–14710. [Google Scholar] [CrossRef] [Green Version]
- Bhattacharya, T.; Newton, I.L.G.; Hardy, R.W. Wolbachia elevates host methyltransferase expression to block an RNA virus early during infection. PLoS Pathog. 2017, 13, e1006427. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Selberg, S.; Blokhina, D.; Aatonen, M.; Koivisto, P.; Siltanen, A.; Mervaala, E.; Kankuri, E.; Karelson, M. Discovery of Small Molecules that Activate RNA Methylation through Cooperative Binding to the METTL3-14-WTAP Complex Active Site. Cell Rep. 2019, 26, 3762–3771.e5. [Google Scholar] [CrossRef] [PubMed] [Green Version]

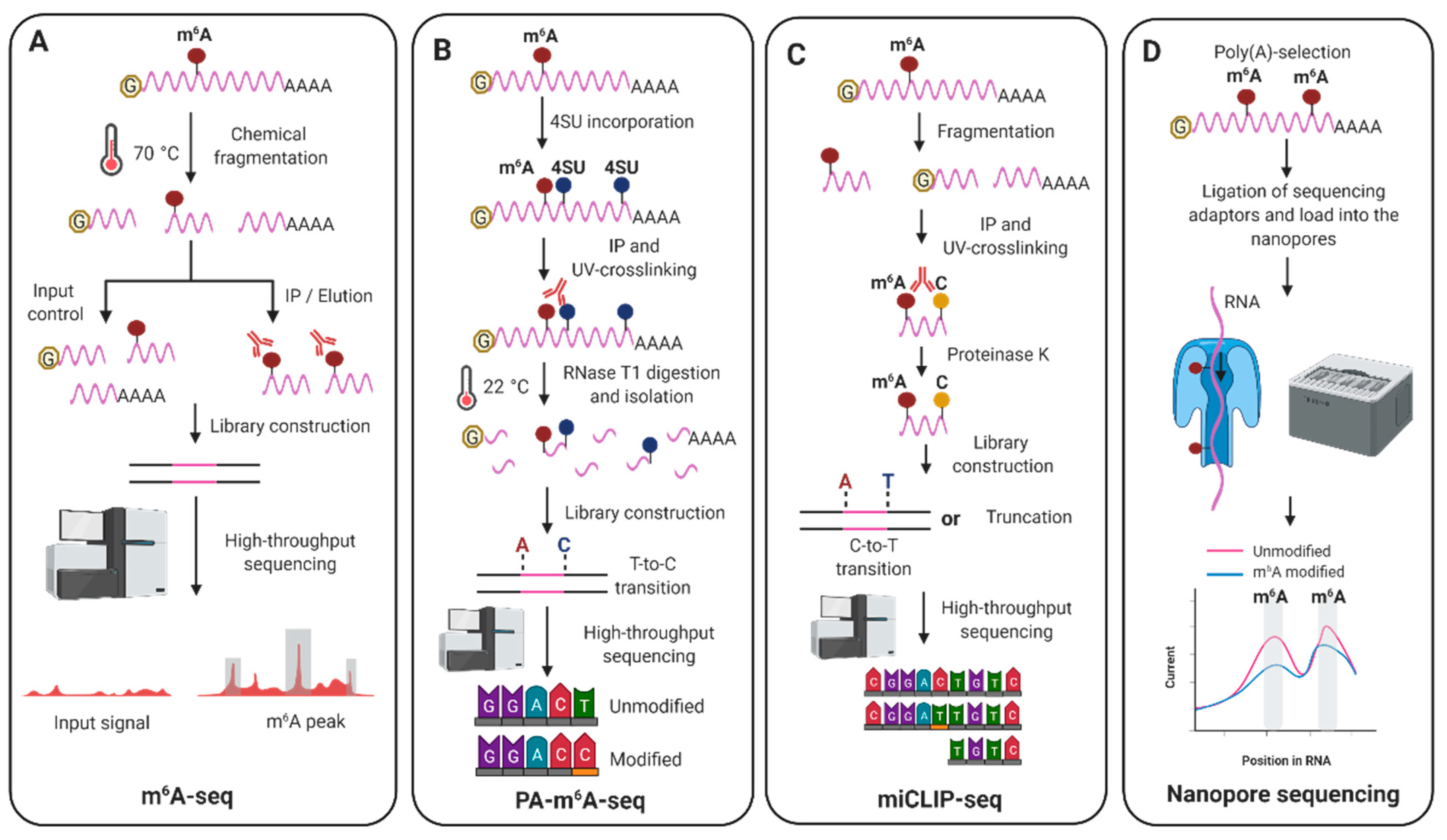

| Virus | Detection | Phenotype | Main Function for m6A | Reference |
|---|---|---|---|---|
| RNA Viruses | ||||
| HIV-1 | m6A-seq | Proviral | vRNA nuclear export | [103] |
| PA-m6A-seq | Proviral | vRNA abundance and protein expression | [104] | |
| m6A-seq | Antiviral | YTHDF1–3 proteins inhibit HIV-1 infection by decreasing HIV-1 reverse transcription | [105,106] | |
| - | Antiviral | YTHDF3 inhibits HIV-1 infection at the step of reverse transcription | [107] | |
| HCV | m6A-seq | Antiviral | m6A modifications in E1 are bound by YTHDF proteins to negatively regulate HCV packaging | [67] |
| ZIKV | m6A-seq | Antiviral | vRNA abundance and protein expression | [68] |
| DENV WNV | m6A-seq | - | - | [67] |
| PEDV | m6A-seq | Antiviral | m6A modifications reduce viral RNA production and viral titers | [108] |
| IAV | PA-m6A-seq | Proviral | m6A modifications increase IAV RNA expression | [109] |
| RSV | m6A-seq | Proviral | m6A modifications enhance RSV replication and gene expression | [70] |
| SARS-CoV2 | m6A-seq and miCLIP | Antiviral | m6A inhibits SARS-CoV2 replication | [110] |
| CHIKV | m6A-IP | Proviral and antiviral | YTHDF1 and YTHDF3 restrict CHIKV replication, while YTHDF2 promotes it | [111] |
| EV71 | m6A-seq | Proviral | METTL3 induces enhanced sumoylation and ubiquitination of the viral RNA polymerase to facilitate viral replication | [112] |
| HMPV | m6A-seq | Proviral | m6A modifications enable vRNA to escape recognition by RIG-I | [113] |
| VSV | miCLIP | Proviral | m6A modifications reduce viral dsRNA formation leading to reduced virus-sensing by innate receptors | [114] |
| DNA VIRUSES | ||||
| SV40 | PA-m6A-seq | Proviral | m6A enhances the translation of viral late transcripts | [115] |
| KSHV | m6A-IP | Proviral | Splicing of ORF50 pre-mRNA | [116] |
| m6A-seq | Antiviral | vRNA stability of latent and lytic transcripts | [117] | |
| m6A-seq | Antiviral and proviral | vRNA abundance and protein expression | [102] | |
| m6A-seq | Proviral | vRNA abundance and protein expression | [59] | |
| EBV | m6A-seq | Both | vRNA stability of latent and lytic transcripts | [118] |
| m6A-seq and PA-m6A-seq | Antiviral | YTHDF1 promotes EBV RNA decay | [119] | |
| Ad5 | m6A-seq and nanopore | Proviral | m6A is required for splicing of adenoviral late transcripts | [93] |
| HBV | m6A-seq | Antiviral and proviral | vRNA stability and viral reverse-transcription | [120] |
| Virus | Depletion/Knockout | Overexpression | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| METTL3 | METTL14 | FTO | ALKBH5 | YTHDF1 | YTHDF2 | YTHDF3 | METTL3 | METTL14 | FTO | ALKBH5 | YTHDF1 | YTHDF2 | YTHDF3 | ||||||||||
| HIV-1 | [103,106] | [103,106] | [106] | [103,106] | [105,106] | [105,106] | [104,105] | [105,106,107] | [104] | [105,106] | [104] | [105,106] | [104] | [105,106] | |||||||||
| HCV | |||||||||||||||||||||||
| ZIKA | |||||||||||||||||||||||
| CHIKV | |||||||||||||||||||||||
| PEDV | |||||||||||||||||||||||
| IAV | [109,121] | [109] | [109] | [109] | |||||||||||||||||||
| RSV | |||||||||||||||||||||||
| SARS-2 | |||||||||||||||||||||||
| EV71 | |||||||||||||||||||||||
| HMPV | |||||||||||||||||||||||
| VSV | [122] | [114] | [122] | [122] | [122] | [123] | [123] | [123] | [114] | [122] | |||||||||||||
| SV40 | |||||||||||||||||||||||
| KSHV | [102] | [59,102,116] | [59,116] | [102,117] | [59] | [102,117] | [59,102] | [102,117] | [59] | [117] | |||||||||||||
| EBV | [119] | [118] | [118,119] | [118] | [119] | [119] | [118] | [118,119] | [119] | ||||||||||||||
| Ad5 | |||||||||||||||||||||||
| HBV | |||||||||||||||||||||||
| HCMV | [121,124] | [121,124] | [121,124] | [121,124] | [121] | [121] | [121] | ||||||||||||||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Baquero-Perez, B.; Geers, D.; Díez, J. From A to m6A: The Emerging Viral Epitranscriptome. Viruses 2021, 13, 1049. https://0-doi-org.brum.beds.ac.uk/10.3390/v13061049
Baquero-Perez B, Geers D, Díez J. From A to m6A: The Emerging Viral Epitranscriptome. Viruses. 2021; 13(6):1049. https://0-doi-org.brum.beds.ac.uk/10.3390/v13061049
Chicago/Turabian StyleBaquero-Perez, Belinda, Daryl Geers, and Juana Díez. 2021. "From A to m6A: The Emerging Viral Epitranscriptome" Viruses 13, no. 6: 1049. https://0-doi-org.brum.beds.ac.uk/10.3390/v13061049






