Fatal Neonatal Sepsis Associated with Human Adenovirus Type 56 Infection: Genomic Analysis of Three Recent Cases Detected in the United States
Abstract
:1. Introduction
2. Materials and Methods
2.1. Case Identification and Clinical Data Abstraction
- Case 1 (Ohio, 2014). An 8-day old, 23-week gestational age, premature female infant developed sepsis, pneumonia, hepatitis, and thrombocytopenia. Human adenovirus DNA was detected in the nasopharynx and blood prompting therapy with cidofovir and intravenous immunoglobulin (IVIG). Despite these interventions, the infant died at 18 days of age. Detailed clinical information for this case was previously reported by Moallem et al. [13]. Human adenovirus DNA was detected in the nasopharynx and blood. The HAdV strain isolated from a nasopharyngeal aspirate, NCH-PRO95, was originally partially characterized at Lovelace Biomedical Research Institute (LBRI) and typed as an intertypic recombinant HAdV-D with a type 15/29 hexon gene (H) and a type 9 fiber gene (F) as reported [13];
- Case 2 (New York, 2019). A 9-day old, full-term male infant experienced fever and poor feeding and was admitted to a local hospital in NY state. The infant was well for the first week of life, and his New York State Newborn Screening was negative for all diseases tested for in the standard screening program at the state newborn screen laboratory (https://www.wadsworth.org/programs/newborn/screening/screened-disorders). Physical examination and initial laboratory studies were nondiagnostic, and intravenous ampicillin, gentamicin, and acyclovir were administered. Bacterial cultures of blood and urine specimens were negative; surface cultures and blood PCR for herpes simplex virus were negative as well. A PCR-based respiratory virus panel test (Panther Fusion, Hologic, Inc., Marlborough, MA, USA) conducted on a nasopharyngeal specimen was positive for both adenovirus and rhinovirus. Because of progressive hypoxia and continued fever, the patient was transferred to a children’s hospital after 72 h of therapy. Further history was obtained that the infant’s mother and grandmother had recent episodes of conjunctivitis around the time of the child’s birth. The physical examination continued to be unremarkable except for temperatures up to 40.2 °C. A chest radiograph showed bilateral diffuse opacities indicative of either infiltrates or atelectasis. Over the subsequent 48 h, he developed worsening tachypnea and oxygen desaturation and was transferred to the pediatric intensive care unit (ICU). Bacterial cultures of blood and urine continued to be negative. His respiratory function rapidly declined, and intubation with mechanical ventilation, followed by provision of inhaled nitric oxide and later extracorporeal membrane oxygenation (ECMO) were required. Serum PCR assays conducted at Eurofins Viracor-IBT laboratories (Lee’s Summit, MO, USA) for enterovirus were negative, but those for HAdV were positive at 1.6 × 109 genome copies/mL. Cidofovir was administered intravenously and ECMO maintained, but the infant had continued respiratory failure and developed hepatorenal failure and coagulopathy. He died at 19 days of age;
- Case 3 (Pennsylvania, 2020). A 14-day old, full-term infant male presented with a 5-day history of poor feeding and new-onset respiratory distress and lethargy. He previously had been noted to have bilateral conjunctival erythema at 6 days of life. Due to the severity of his illness, he was taken to an outside emergency department, where he was noted to have severe retractions and multiple apneic events. He was intubated but continued to have hypoxemia despite escalation to high-frequency oscillatory ventilation, so he was transferred to a children’s hospital where he was cannulated onto ECMO. He required significant vasopressor support and also had renal failure requiring renal replacement therapy. Cerebrospinal fluid studies were unremarkable. He was given broad spectrum antimicrobials, including vancomycin, cefepime, and acyclovir. Bacterial cultures were negative. An in-house respiratory virus quantitative real-time PCR panel [14] was positive for HAdV. A serum HAdV PCR also tested positive. Due to the severity of his presentation, IVIG was given daily for 2 days. Cidofovir was held due to the concerns for renal toxicity. Because he continued to have clinical instability and repeat serum HAdV PCR testing 3 days later was positive, he was given cidofovir dosed at 5 mg/kg with hyperhydration. Over the following weeks, serial serum HAdV PCR testing remained positive, and he received 2 additional doses of cidofovir on a weekly basis. He also received 5 additional doses of IVIG. Despite these interventions, there was little change in his clinical status, and he continued to require ECMO support due to the fact of respiratory failure and severe hemodynamic instability. On day 42 of life, he was noted to have worsening leukopenia and thrombocytopenia with associated hemodynamic instability. He then developed both pulmonary and gastrointestinal hemorrhage. Due to the bleeding, he was decannulated from ECMO and died on day 43 of life.
2.2. Virus Isolation and Initial Molecular Typing
2.3. Whole Genome Sequencing and Annotation
2.4. Genomic Analyses
3. Results
3.1. Preliminary Virus Typing
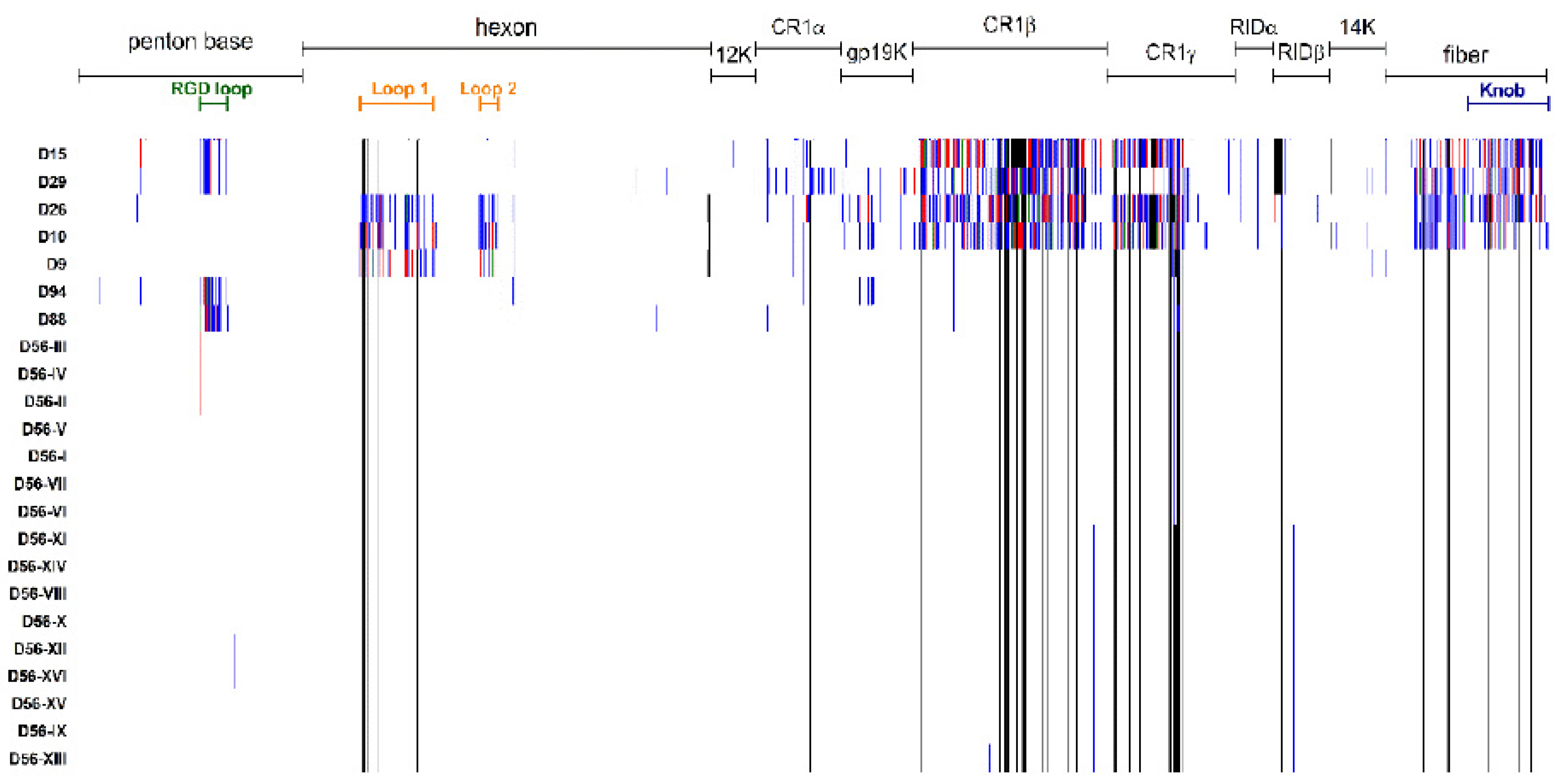
3.2. Genetic Analysis of Virus Isolates
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Green, W.R.; Williams, A.W. Neonatal adenovirus pneumonia. Arch. Pathol. Lab. Med. 1989, 113, 190–191. [Google Scholar] [PubMed]
- Ronchi, A.; Doern, C.; Brock, E.; Pugni, L.; Sánchez, P.J. Neonatal adenoviral infection: A seventeen year experience and review of the literature. J. Pediatr. 2014, 164, 529–535.e4. [Google Scholar] [CrossRef] [PubMed]
- Abzug, M.J.; Levin, M.J. Neonatal adenovirus infection: Four patients and review of the literature. Pediatrics 1991, 87, 890–896. [Google Scholar]
- Sun, C.C.; Duara, S. Fatal adenovirus pneumonia in two newborn infants, one case caused by adenovirus type 30. Pediatr. Pathol. 1985, 4, 247–255. [Google Scholar] [CrossRef] [PubMed]
- De Jong, J.C.; Wermenbol, A.G.; Verweij-Uijterwaal, M.W.; Slaterus, K.W.; Wertheim-Van Dillen, P.; Van Doornum, G.J.; Khoo, S.H.; Hierholzer, J.C. Adenoviruses from human immunodeficiency virus-infected individuals, including two strains that represent new candidate serotypes Ad50 and Ad51 of species B1 and D, respectively. J. Clin. Microbiol. 1999, 37, 3940–3945. [Google Scholar] [CrossRef] [Green Version]
- Robinson, C.M.; Singh, G.; Henquell, C.; Walsh, M.P.; Peigue-Lafeuille, H.; Seto, D.; Jones, M.S.; Dyer, D.W.; Chodosh, J. Computational analysis and identification of an emergent human adenovirus pathogen implicated in a respiratory fatality. Virology 2011, 409, 141–147. [Google Scholar] [CrossRef] [PubMed]
- Henquell, C.; Boeuf, B.; Mirand, A.; Bacher, C.; Traore, O.; Déchelotte, P.; Labbé, A.; Bailly, J.L.; Peigue-Lafeuille, H. Fatal adenovirus infection in a neonate and transmission to health-care workers. J. Clin. Virol. 2009, 45, 345–348. [Google Scholar] [CrossRef]
- Gonzalez, G.; Yawata, N.; Aoki, K.; Kitaichi, N. Challenges in management of epidemic keratoconjunctivitis with emerging recombinant human adenoviruses. J. Clin. Virol. 2019, 112, 1–9. [Google Scholar] [CrossRef]
- Huang, G.; Yao, W.; Yu, W.; Mao, L.; Sun, H.; Yao, W.; Tian, J.; Wang, L.; Bo, Z.; Zhu, Z.; et al. Outbreak of epidemic keratoconjunctivitis caused by human adenovirus type 56, China, 2012. PLoS ONE 2014, 9, e110781. [Google Scholar] [CrossRef] [PubMed]
- Hiroi, S.; Furubayashi, K.; Kawahata, T.; Morikawa, S.; Kase, T. A case of urethritis caused by human adenovirus type 56. Jpn. J. Infect. Dis. 2012, 65, 273–274. [Google Scholar] [CrossRef] [Green Version]
- Idigoras, P.; Zapico, M.S.; Montes, M.; Romano, C. Simultaneous urethritis and conjunctivitis caused by adenovirus type 56. Med. Clin. 2014, 142, 558. [Google Scholar] [CrossRef] [PubMed]
- Hanaoka, N.; Ito, S.; Konagaya, M.; Nojiri, N.; Yasuda, M.; Fujimoto, T.; Deguchi, T. Infectious human adenoviruses are shed in urine even after disappearance of urethral symptoms. PLoS ONE 2019, 14, e0212434. [Google Scholar] [CrossRef] [Green Version]
- Moallem, M.; Song, E.; Jaggi, P.; Conces, M.R.; Kajon, A.E.; Sánchez, P.J. Adenovirus and “Culture-Negative Sepsis” in a Preterm Neonate. AJP Rep. 2016, 6, e417–e420. [Google Scholar]
- Kajon, A.E.; Lamson, D.M.; Spiridakis, E.; Cardenas, A.M.; Babady, N.E.; Fisher, B.T.; St George, K. Isolation of a novel intertypic recombinant human mastadenovirus B2 from two unrelated bone marrow transplant recipients. New Microbes New Infect. 2020, 35, 100677. [Google Scholar] [CrossRef]
- Lamson, D.M.; Kajon, A.; Shudt, M.; Girouard, G.; St George, K. Detection and Genetic Characterization of Adenovirus Type 14 Strain in Students with Influenza-Like Illness, New York, USA, 2014–2015. Emerg Infect. Dis. 2017, 23, 1194–1197. [Google Scholar] [CrossRef]
- Shean, R.C.; Makhsous, N.; Stoddard, G.D.; Lin, M.J.; Greninger, A.L. VAPiD: A lightweight cross-platform viral annotation pipeline and identification tool to facilitate virus genome submissions to NCBI GenBank. BMC Bioinform. 2019, 20, 48. [Google Scholar] [CrossRef] [Green Version]
- Johnson, M.; Zaretskaya, I.; Raytselis, Y.; Merezhuk, Y.; McGinnis, S.; Madden, T.L. NCBI BLAST: A better web interface. Nucleic Acids Res. 2008, 36, W5–W9. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [Green Version]
- Edler, D.; Klein, J.; Antonelli, A.; Silvestro, D. raxmlGUI 2.0 beta: A graphical interface and toolkit for phylogenetic analyses using RAxML. BioRxiv 2019, 800912. [Google Scholar] [CrossRef]
- Lole, K.S.; Bollinger, R.C.; Paranjape, R.S.; Gadkari, D.; Kulkarni, S.S.; Novak, N.G.; Ingersoll, R.; Sheppard, H.W.; Ray, S.C. Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J. Virol. 1999, 73, 152–160. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fox, J. Getting started with the R commander: A basic-statistics graphical user interface to R. J. Stat. Softw. 2005, 14, 1–42. [Google Scholar] [CrossRef] [Green Version]
- Obenauer, J.C.; Denson, J.; Mehta, P.K.; Su, X.; Mukatira, S.; Finkelstein, D.B.; Xu, X.; Wang, J.; Ma, J.; Fan, Y.; et al. Large-scale sequence analysis of avian influenza isolates. Science 2006, 311, 1576–1580. [Google Scholar] [CrossRef]
- Kaneko, H.; Aoki, K.; Ohno, S.; Ishiko, H.; Fujimoto, T.; Kikuchi, M.; Harada, S.; Gonzalez, G.; Koyanagi, K.O.; Watanabe, H. Complete genome analysis of a novel intertypic recombinant human adenovirus causing epidemic keratoconjunctivitis in Japan. J. Clin. Microbiol. 2011, 49, 484–490. [Google Scholar] [CrossRef] [Green Version]
- Laverty, C.R.; Russell, P.; Black, J.; Kappagoda, N.; Booth, N. Adenovirus infection of the cervix. Acta Cytol. 1977, 21, 114–117. [Google Scholar]
- Harnett, G.B.; Newnham, W.A. Isolation of adenovirus type 19 from the male and female genital tracts. Br. J. Vener. Dis. 1981, 57, 55–57. [Google Scholar] [CrossRef] [Green Version]
- Phillips, P.A.; Harnett, G.B.; Gollow, M.M. Adenovirus type 19 and a closely related new serotype in genital infection. Br. J. Vener. Dis. 1982, 58, 131–132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Montone, K.T.; Furth, E.E.; Pietra, G.G.; Gupta, P.K. Neonatal adenovirus infection: A case report with in situ hybridization confirmation of ascending intrauterine infection. Diagn. Cytopathol. 1995, 12, 341–344. [Google Scholar] [CrossRef]
- Ornelles, D.A.; Gooding, L.R.; Garnett-Benson, C. Neonatal infection with species C adenoviruses confirmed in viable cord blood lymphocytes. PLoS ONE 2015, 10, e0119256. [Google Scholar] [CrossRef] [Green Version]
- Tsekoura, E.A.; Konstantinidou, A.; Papadopoulou, S.; Athanasiou, S.; Spanakis, N.; Kafetzis, D.; Antsaklis, A.; Tsakris, A. Adenovirus genome in the placenta: Association with histological chorioamnionitis and preterm birth. J. Med. Virol. 2010, 82, 1379–1383. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schaap, G.J.; de Jong, J.C.; van Bijsterveld, O.P.; Beekhuis, W.H. A new intermediate adenovirus type causing conjunctivitis. Arch. Ophthalmol. 1979, 97, 2336–2338. [Google Scholar] [CrossRef]
- Liddle, O.L.; Samuel, M.I.; Sudhanva, M.; Ellis, J.; Taylor, C. Adenovirus urethritis and concurrent conjunctivitis: A case series and review of the literature. Sex. Transm. Infect. 2015, 91, 87–90. [Google Scholar] [CrossRef]
- Hiroi, S.; Kawahata, T.; Furubayashi, K. First isolation of human adenovirus type 85 by molecular analysis of adenoviruses in cases of urethritis. J. Med. Microbiol. 2020, 69, 265–269. [Google Scholar] [CrossRef] [PubMed]
- Samaraweera, G.R.; Garcia, K.; Druce, J.; Williams, H.; Bradshaw, C.S.; Fairley, C.K.; Chow, E.P.; Denham, I.M.; Read, T.R.; Chen, M.Y. Characteristics of adenovirus urethritis among heterosexual men and men who have sex with men: A review of clinical cases. Sex. Transm. Infect. 2016, 92, 172–174. [Google Scholar] [CrossRef] [PubMed]
- Adrian, T.; Bastian, B.; Benoist, W.; Hierholzer, J.C.; Wigand, R. Characterization of adenovirus 15/H9 intermediate strains. Intervirology 1985, 23, 15–22. [Google Scholar] [CrossRef] [PubMed]
- Adrian, T.; Bastian, B.; Wagner, V. Restriction site mapping of adenovirus types 9 and 15 and genome types of intermediate adenovirus 15/H9. Intervirology 1989, 30, 169–176. [Google Scholar] [CrossRef] [PubMed]
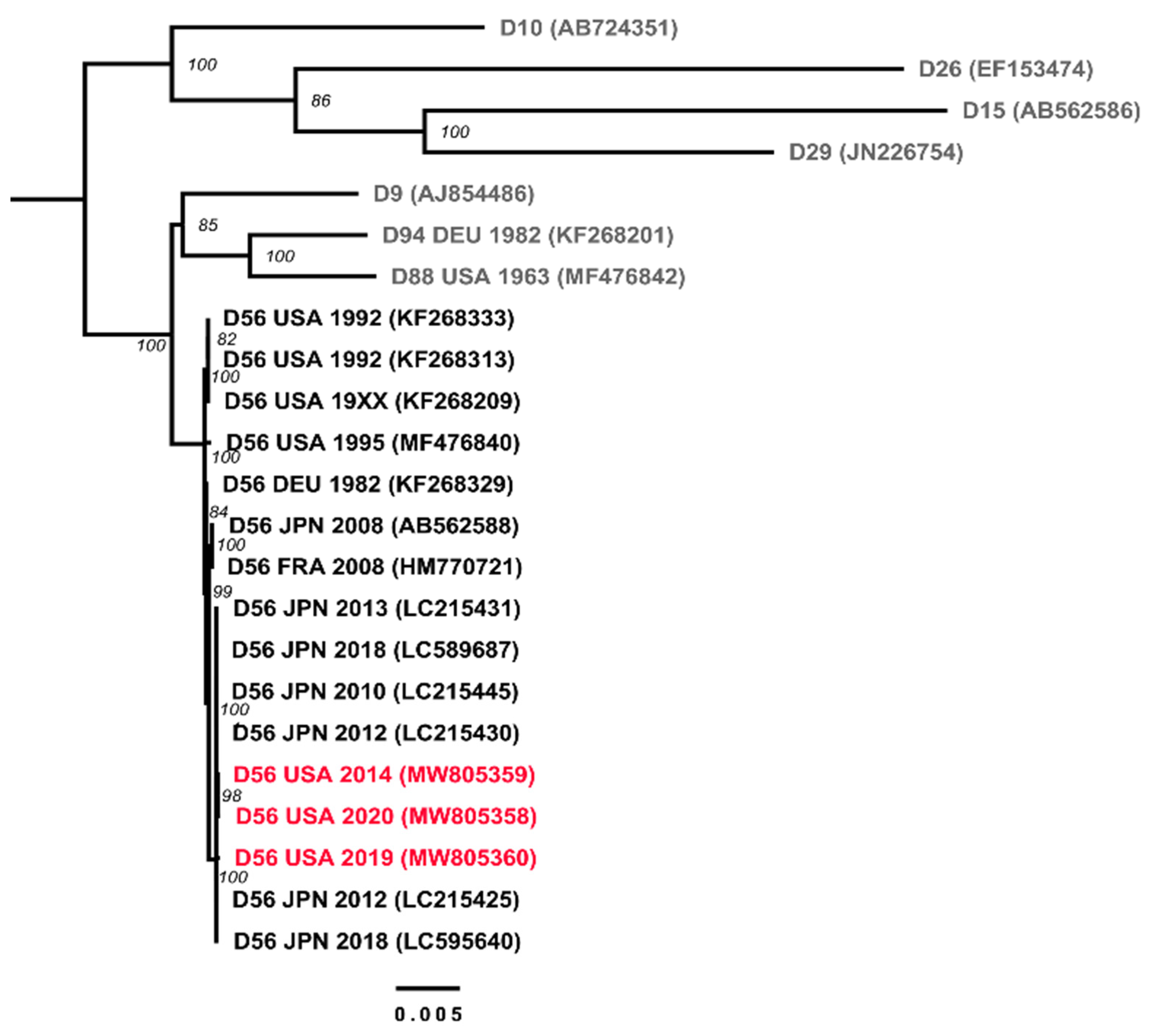
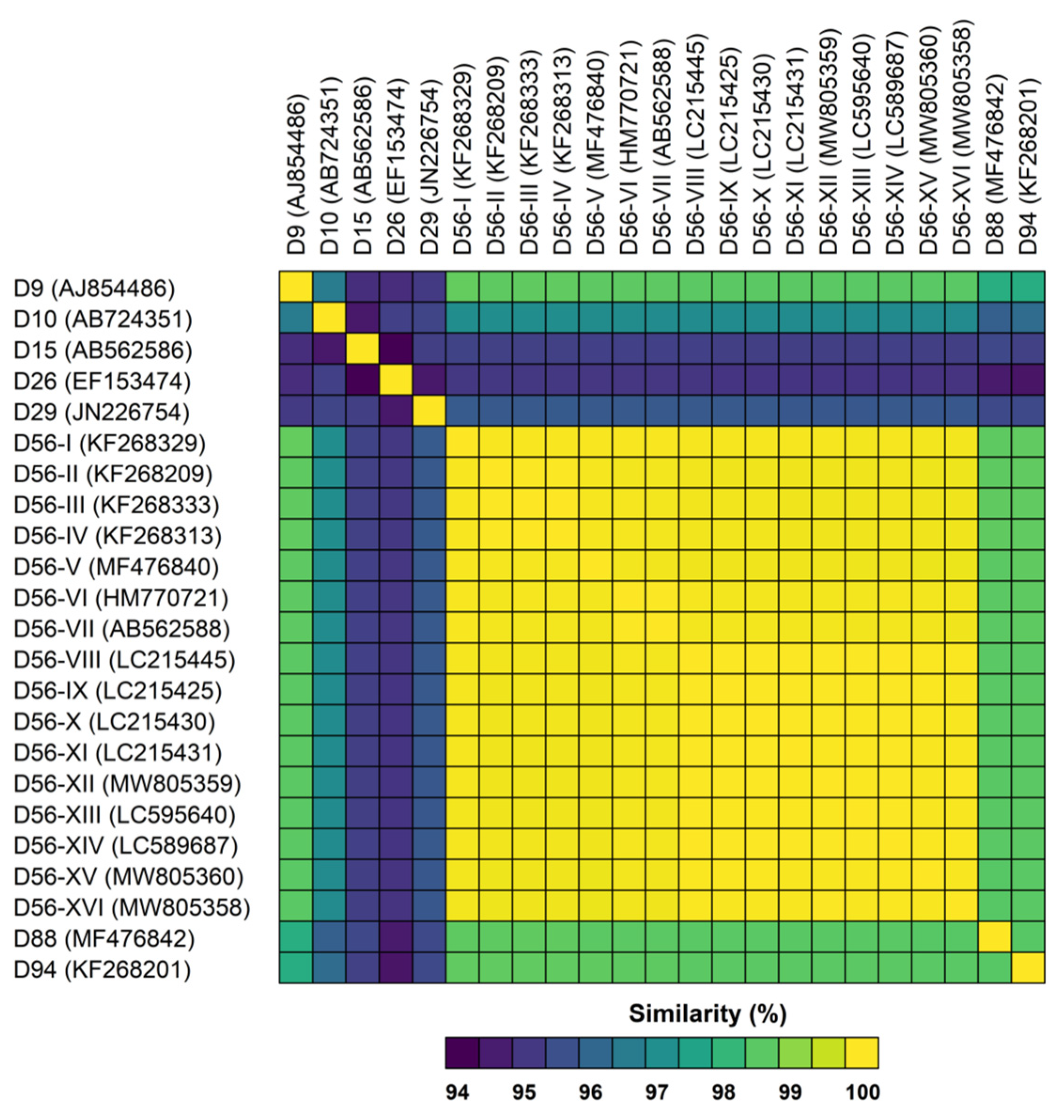
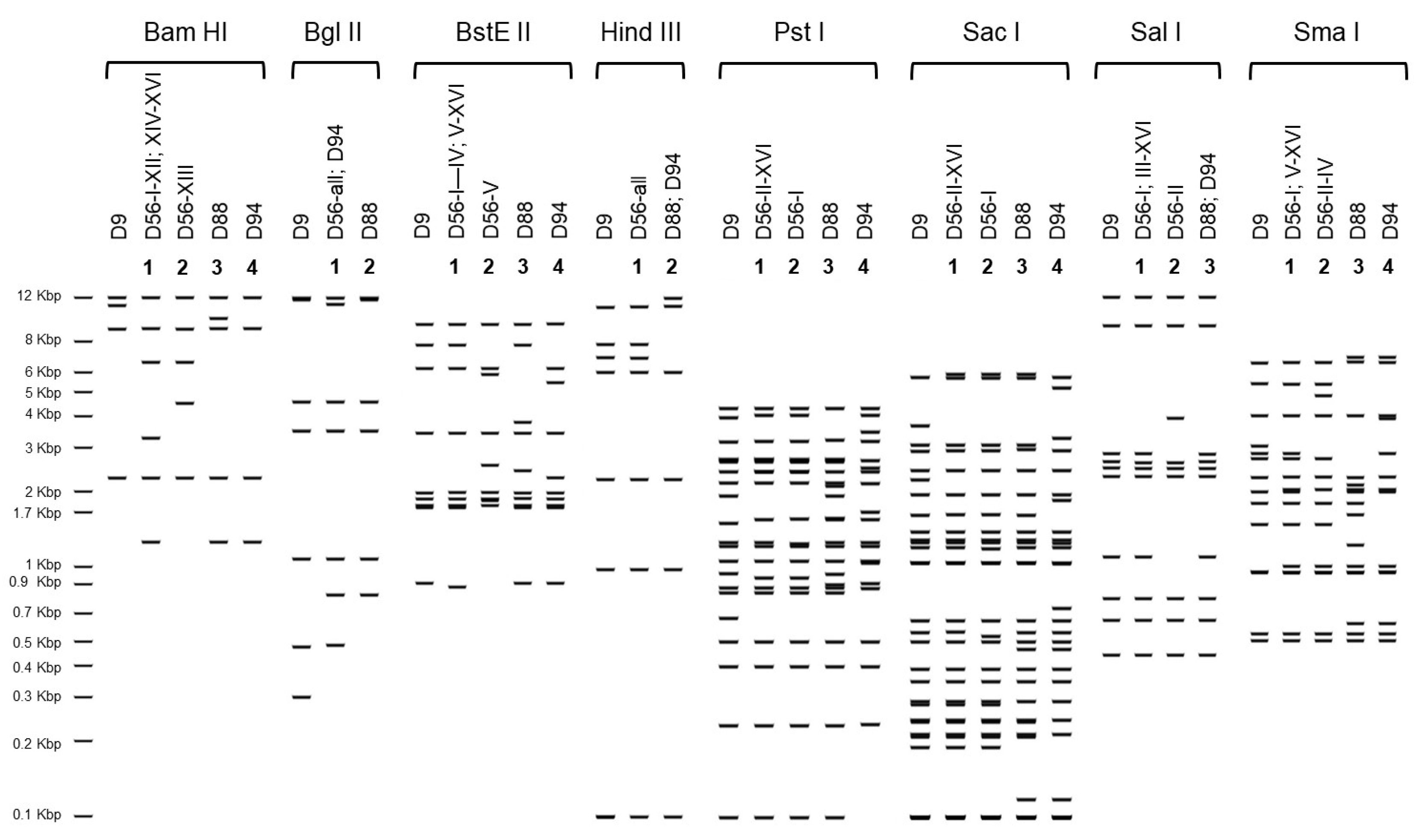
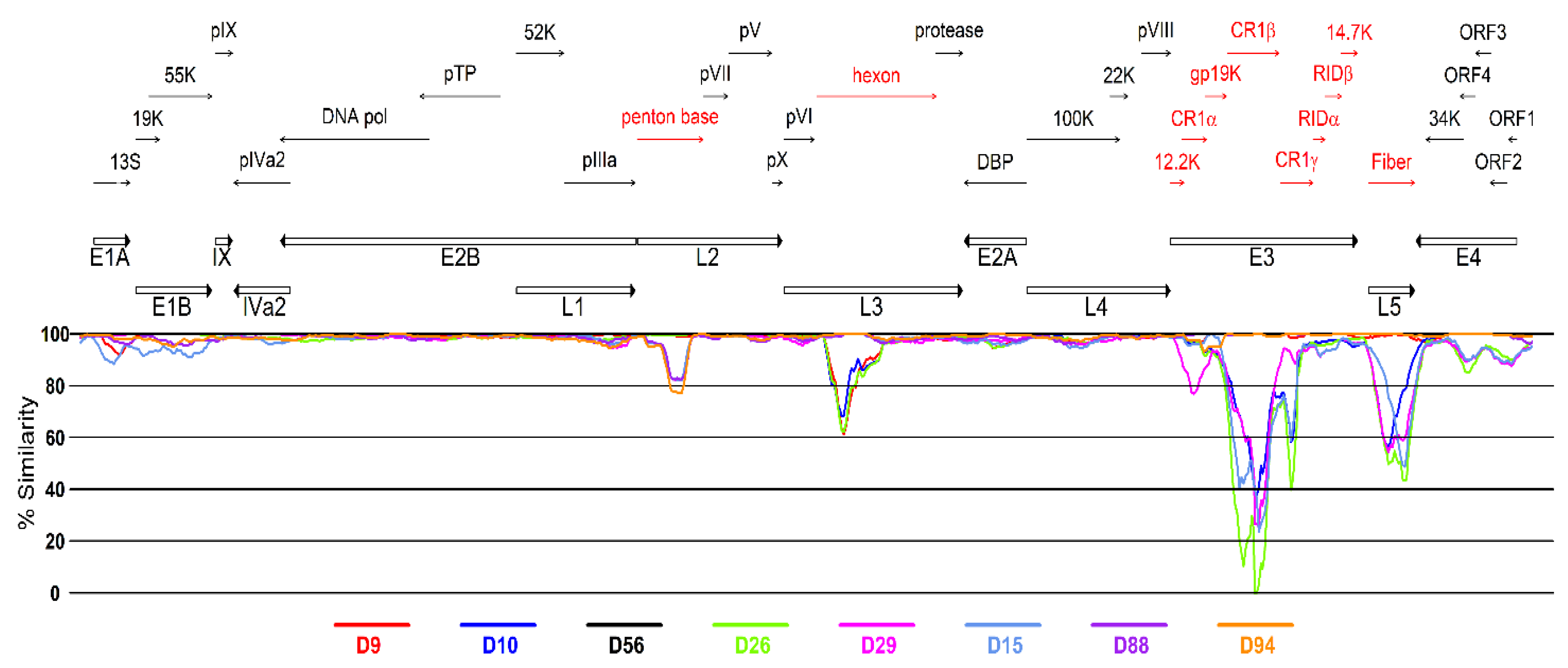
| Strain Designation | Genotype | Assigned ID for This Study | Source | Clinical Illness | Year of Isolation | Place of Isolation | Accession No. | Molecular Type | ||
|---|---|---|---|---|---|---|---|---|---|---|
| P | H | F | ||||||||
| Hicks (p) | D9 | D9 | Anal specimen | Arthritis | 1954 | MA, USA | AJ854486 | 9 | 9 | 9 |
| J.J. (p) | D10 | D10 | Conjunctival swab | Conjunctivitis | 195X | D.C., USA | AB724351 | 10 | 10 | 10 |
| CH38 (p) | D15 | D15 | Conjunctival scrapings | Conjunctivitis | 1955 | SAU | AB562586 | 15 | 15 | 15 |
| BP-2 (p) | D26 | D26 | Anal specimen | None | 1956 | D.C., USA | EF153474 | 26 | 26 | 26 |
| BP-6 (p) | D29 | D29 | Anal specimen | NA | 1959 | D.C., USA | JN226754 | 29 | 15 | 29 |
| MEE-MOLD | D88 | D88 | Respiratory specimen | NA | 1963 | MA, USA | MF476842 | 88 | 15 | 9 |
| HEIM 00080 | D94 | D94 | NA | NA | 1982 | DEU | KF268201 | 33 | 15 | 9 |
| HEIM 00081 | D56 | D56-I | NA | NA | 1982 | DEU | KF268329 | 9 | 15 | 9 |
| MEEI 00078 | D56 | D56-II | NA | NA | 19XX | MA, USA | KF268209 | 9 | 15 | 9 |
| CL 50 | D56 | D56-III | NA | NA | 1992 | PA, USA | KF268333 | 9 | 15 | 9 |
| Pitts 00150 | D56 | D56-IV | NA | NA | 1992 | PA, USA | KF268313 +++ | 9 | 15 | 9 |
| MEE-CHBR | D56 | D56-V | Vaginal swab | NA | 1995 | MA, USA | MF476840 | 9 | 15 | 9 |
| p | D56 | D56-VI | Lung biopsy | Sepsis N | 2008 | FRA | HM770721 | 9 | 15 | 9 |
| 2307-S | D56 | D56-VII | Conjunctival swab | EKC | 2008 | Sapporo, JPN | AB562588 | 9 | 15 | 9 |
| 20101537 | D56 | D56-VIII | Conjunctival swab | EKC | 2010 | Kumamoto, JPN | LC215445 | 9 | 15 | 9 |
| 20121516 | D56 | D56-IX | Conjunctival swab | EKC | 2012 | Kumamoto, JPN | LC215425 | 9 | 15 | 9 |
| 20121569 | D56 | D56-X | Conjunctival swab | EKC | 2012 | Kumamoto, JPN | LC215430 | 9 | 15 | 9 |
| 20131505 | D56 | D56-XI | Conjunctival swab | EKC | 2013 | Kumamoto, JPN | LC215431 | 9 | 15 | 9 |
| NCH-Pro95 | D56 | D56-XII | Respiratory specimen | Sepsis N | 2014 | OH, USA | MW805359 § | 9 | 15 | 9 |
| TKY3113Asew | D56 | D56-XIII | Sewage water | --- | 2018 | Tokyo, JPN | LC595640 | 9 | 15 | 9 |
| TKYAd188507 | D56 | D56-XIV | Urine | Urethritis | 2018 | Tokyo, JPN | LC589687 | 9 | 15 | 9 |
| IDR1900044114 | D56 | D56-XV | Respiratory specimen | Sepsis N | 2019 | NY, USA | MW805360 § | 9 | 15 | 9 |
| VIR209329 | D56 | D56-XVI | Respiratory specimen | Sepsis N | 2020 | PA, USA | MW805358 § | 9 | 15 | 9 |
| Nucleotide Position § | Affected Amino Acid Position ¥ | Gene | Protein Product | D56-VI (HM770721) | D56-XII (MW805359) | D56-XV (MW805360) | D56-XVI (MW805358) | |||
|---|---|---|---|---|---|---|---|---|---|---|
| AA | Depth ჶ | AA | Depth ჶ | AA | Depth ჶ | |||||
| 788 | 217 | E1a | 12S | L | I | 25 | I | 28 | I | 33 |
| 1090 | 519 | 13S | M | M | 36 | I | 25 | M | 35 | |
| 1971 | 398 | E1B | 19K | R | K | 33 | K | 24 | K | 24 |
| 10992 | 383 | L1 | 52/55K | S | N | 18 | N | 21 | N | 28 |
| 14397 | 913 | L2 | penton base | D | N | 28 | N | 21 | N | 33 |
| 14572 | 1088 | penton base | P | L | 33 | P | 16 | L | 48 | |
| 15862 | 197 | pV | H | R | 45 | R | 15 | R | 35 | |
| 28582 | 1075 | E3 | CR1β | E | K | 35 | K | 20 | K | 29 |
| 28648 | 1141 | CR1β | I | F | 29 | F | 20 | F | 33 | |
| 29991 | 140 | 14.5K | V | A | 29 | A | 15 | A | 39 | |
| 31837 | 943 | L5 | fiber | D | Y | 29 | Y | 16 | Y | 24 |
| 33085 | 346 | E4 | Orf 4 | S | P | 23 | P | 21 | P | 31 |
| 33119 | 20 | 34K | S | C | 20 | S | 23 | C | 27 | |
| 33119 | 312 | Orf 4 | I | M | 20 | I | 23 | M | 27 | |
| 34405 | 189 | Orf 1 | L | L | 28 | F | 21 | L | 28 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Otto, W.R.; Lamson, D.M.; Gonzalez, G.; Weinberg, G.A.; Pecora, N.D.; Fisher, B.T.; St. George, K.; Kajon, A.E. Fatal Neonatal Sepsis Associated with Human Adenovirus Type 56 Infection: Genomic Analysis of Three Recent Cases Detected in the United States. Viruses 2021, 13, 1105. https://0-doi-org.brum.beds.ac.uk/10.3390/v13061105
Otto WR, Lamson DM, Gonzalez G, Weinberg GA, Pecora ND, Fisher BT, St. George K, Kajon AE. Fatal Neonatal Sepsis Associated with Human Adenovirus Type 56 Infection: Genomic Analysis of Three Recent Cases Detected in the United States. Viruses. 2021; 13(6):1105. https://0-doi-org.brum.beds.ac.uk/10.3390/v13061105
Chicago/Turabian StyleOtto, William R., Daryl M. Lamson, Gabriel Gonzalez, Geoffrey A. Weinberg, Nicole D. Pecora, Brian T. Fisher, Kirsten St. George, and Adriana E. Kajon. 2021. "Fatal Neonatal Sepsis Associated with Human Adenovirus Type 56 Infection: Genomic Analysis of Three Recent Cases Detected in the United States" Viruses 13, no. 6: 1105. https://0-doi-org.brum.beds.ac.uk/10.3390/v13061105







