Development of a Bio-Layer Interferometry-Based Protease Assay Using HIV-1 Protease as a Model
Abstract
:1. Introduction
2. Materials and Methods
2.1. Expression and Purification of HIV-1 PR
2.2. Cloning, Expression and Purification of Recombinant Fluorescent Substrates
2.3. Preparation and Investigation of Ni2+-, Cu2+-, Zn2+- and Co2+-Charged NTA Biosensors
2.4. Real-Time HIV-1 PR Activity Measurements by BLI Using Purified Substrates
2.5. BLI Protease Assay Using Non-Purified Substrate
2.6. PAGE Analysis and Densitometry
2.7. Enzyme Kinetic Measurements
3. Results
3.1. Recombinants Substrates
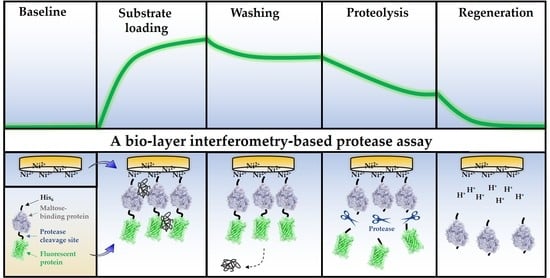
3.2. Design of the BLI-Based Protease Assay
3.3. Optimizing Substrate Immobilization on Biosensors
3.4. The Effect of Enzyme Concentration on Substrate Processing
3.5. Measuring the Effect of an Inhibitor on Protease Activity
3.6. Effect of P2-Leu Substitution in HIV-1 PR MA/CA Cleavage Site
3.7. Study on the Potential Substrate Groove of HIV-1 PR
3.8. BLI Protease Assay Using Non-Purified Substrate
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mótyán, J.A.; Tóth, F.; Tőzsér, J. Research applications of proteolytic enzymes in molecular biology. Biomolecules 2013, 3, 923–942. [Google Scholar] [CrossRef] [Green Version]
- Callahan, B.P.; Stanger, M.J.; Belfort, M. Protease Activation of Split Green Fluorescent Protein. Chem. Eur. J. 2010, 11, 2259–2263. [Google Scholar] [CrossRef]
- Nicholls, S.B.; Chu, J.; Abbruzzese, G.; Tremblay, K.D.; Hardy, J.A. Mechanism of a genetically encoded dark-to-bright reporter for caspase activity. J. Biol. Chem. 2011, 286, 24977–24986. [Google Scholar] [CrossRef] [Green Version]
- Mahdi, M.; Mótyán, J.A.; Szojka, Z.I.; Golda, M.; Miczi, M.; Tőzsér, J. Analysis of the efficacy of HIV protease inhibitors against SARS-CoV-2′s main protease. Virol. J. 2020, 17, 190. [Google Scholar] [CrossRef]
- Froggatt, H.M.; Heaton, B.E.; Heaton, N.S. Development of a Fluorescence-Based, High-Throughput SARS-CoV-2 3CLpro Reporter Assay. J. Virol. 2020, 94, e01265-20. [Google Scholar] [CrossRef]
- Kapust, R.B.; Tözsér, J.; Copeland, T.D.; Waugh, D.S. The P1′ specificity of tobacco etch virus protease. Biochem. Biophys. Res. Commun. 2002, 294, 949–955. [Google Scholar] [CrossRef] [Green Version]
- Renicke, C.; Spadaccini, R.; Taxis, C. A tobacco etch virus protease with increased substrate tolerance at the P1′ position. PLoS ONE 2013, 8, e67915. [Google Scholar] [CrossRef] [Green Version]
- Patel, D.; Frelinger, J.; Goudsmit, J.; Kim, B. In Vitro Assay for Site-Specific Proteases Using Bead-Attached GFP Substrate. BioTechniques 2001, 31, 1194–1203. [Google Scholar] [CrossRef]
- Askin, S.P.; Morin, I.; Schaeffer, P.M. Development of a protease activity assay using heat-sensitive Tus–GFP fusion protein substrates. Anal. Biochem. 2011, 415, 126–133. [Google Scholar] [CrossRef]
- Chaparro-Riggers, J.F.; Breves, R.; Michels, A.; Maurer, K.-H.; Bornscheuer, U. A GFP-based assay for the determination of hydrolytic activity and substrate specificity of subtilisins under washing conditions. J. Mol. Catal. B Enzym. 2005, 35, 74–77. [Google Scholar] [CrossRef]
- Gammon, S.T.; Villalobos, V.M.; Roshal, M.; Samrakandi, M.; Piwnica-Worms, D. Rational design of novel red-shifted BRET pairs: Platforms for real-time single-chain protease biosensors. Biotechnol. Prog. 2009, 25, 559–569. [Google Scholar] [CrossRef] [Green Version]
- Branchini, B.R.; Rosenberg, J.C.; Ablamsky, D.M.; Taylor, K.P.; Southworth, T.L.; Linder, S.J. Sequential bioluminescence resonance energy transfer–fluorescence resonance energy transfer-based ratiometric protease assays with fusion proteins of firefly luciferase and red fluorescent protein. Anal. Biochem. 2011, 414, 239–245. [Google Scholar] [CrossRef] [PubMed]
- Bozóki, B.; Gazda, L.; Tóth, F.; Miczi, M.; Mótyán, J.A.; Tőzsér, J. A recombinant fusion protein-based, fluorescent protease assay for high throughput-compatible substrate screening. Anal. Biochem. 2018, 540–541, 52–63. [Google Scholar] [CrossRef] [PubMed]
- Mótyán, J.A.; Miczi, M.; Bozóki, B.; Tőzsér, J. Data supporting Ni-NTA magnetic bead-based fluorescent protease assay using recombinant fusion protein substrates. Data Brief 2018, 18, 203–208. [Google Scholar] [CrossRef]
- Bozóki, B.; Mótyán, J.A.; Miczi, M.; Gazda, L.D.; Tőzsér, J. Use of Recombinant Fusion Proteins in a Fluorescent Protease Assay Platform and Their In-gel Renaturation. J. Vis. Exp. 2019, e58824. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Golda, M.; Mótyán, J.A.; Mahdi, M.; Tőzsér, J. Functional Study of the Retrotransposon-Derived Human PEG10 Protease. Int. J. Mol. Sci. 2020, 21, 2424. [Google Scholar] [CrossRef] [Green Version]
- Gazda, L.D.; Matúz, K.J.; Nagy, T.; Mótyán, J.A.; Tőzsér, J. Biochemical characterization of Ty1 retrotransposon protease. PLoS ONE 2020, 15, e0227062. [Google Scholar] [CrossRef]
- Miczi, M.; Golda, M.; Kunkli, B.; Nagy, T.; Tőzsér, J.; Mótyán, J.A. Identification of Host Cellular Protein Substrates of SARS-COV-2 Main Protease. Int. J. Mol. Sci. 2020, 21, 9523. [Google Scholar] [CrossRef] [PubMed]
- Bozóki, B.; Mótyán, J.A.; Hoffka, G.; Waugh, D.S.; Tőzsér, J. Specificity Studies of the Venezuelan Equine Encephalitis Virus Non-Structural Protein 2 Protease Using Recombinant Fluorescent Substrates. Int. J. Mol. Sci. 2020, 21, 7686. [Google Scholar] [CrossRef]
- Concepcion, J.; Witte, K.; Wartchow, C.; Choo, S.; Yao, D.; Persson, H.; Wei, J.; Li, P.; Heidecker, B.; Ma, W.; et al. Label-free detection of biomolecular interactions using BioLayer interferometry for kinetic characterization. Comb. Chem. High Throughput Screen. 2009, 12, 791–800. [Google Scholar] [CrossRef] [PubMed]
- Kojima, T.; Nakane, A.; Zhu, B.; Alfi, A.; Nakano, H. A simple, real-time assay of horseradish peroxidase using biolayer interferometry. Biosci. Biotechnol. Biochem. 2019, 83, 1822–1828. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Wilkens, S. Biolayer interferometry of lipid nanodisc-reconstituted yeast vacuolar H+ -ATPase. Protein Sci. 2017, 26, 1070–1079. [Google Scholar] [CrossRef] [Green Version]
- Steinrücke, P.; Aldinger, U.; Hill, O.; Hillisch, A.; Basch, R.; Diekmann, S. Design of helical proteins for real-time endoprotease assays. Anal. Biochem. 2000, 286, 26–34. [Google Scholar] [CrossRef]
- Ong, I.L.H.; Yang, K.-L. Recent developments in protease activity assays and sensors. Analyst 2017, 142, 1867–1881. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tözsér, J.; Oroszlan, S. Proteolytic events of HIV-1 replication as targets for therapeutic intervention. Curr. Pharm. Des. 2003, 9, 1803–1815. [Google Scholar] [CrossRef] [PubMed]
- Konvalinka, J.; Kräusslich, H.-G.; Müller, B. Retroviral proteases and their roles in virion maturation. Virology 2015, 479-480, 403–417. [Google Scholar] [CrossRef] [Green Version]
- Weber, I.; Wang, Y.-F.; Harrison, R.W. HIV Protease: Historical Perspective and Current Research. Viruses 2021, 13, 839. [Google Scholar] [CrossRef]
- Rein, A. Across the Hall from Pioneers. Viruses 2021, 13, 491. [Google Scholar] [CrossRef] [PubMed]
- Gilden, R. Steve Oroszlan: A Personal Perspective. Viruses 2021, 13, 622. [Google Scholar] [CrossRef]
- Tözsér, J. Comparative studies on retroviral proteases: Substrate specificity. Viruses 2010, 2, 147–165. [Google Scholar] [CrossRef] [Green Version]
- Mahalingam, B.; Louis, J.M.; Reed, C.C.; Adomat, J.M.; Krouse, J.; Wang, Y.F.; Harrison, R.W.; Weber, I.T. Structural and kinetic analysis of drug resistant mutants of HIV-1 protease. Eur. J. Biochem. 1999, 263, 238–245. [Google Scholar] [CrossRef] [PubMed]
- Mahalingam, B.; Louis, J.M.; Hung, J.; Harrison, R.W.; Weber, I.T. Structural implications of drug-resistant mutants of HIV-1 protease: High-resolution crystal structures of the mutant protease/substrate analogue complexes. Proteins 2001, 43, 455–464. [Google Scholar] [CrossRef] [PubMed]
- Miklóssy, G.; Tözsér, J.; Kádas, J.; Ishima, R.; Louis, J.M.; Bagossi, P. Novel macromolecular inhibitors of human immunodeficiency virus-1 protease. Protein Eng. Des. Sel. 2008, 21, 453–461. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sahin-Tóth, M. Human cationic trypsinogen. Role of Asn-21 in zymogen activation and implications in hereditary pancreatitis. J. Biol. Chem. 2000, 275, 22750–22755. [Google Scholar] [CrossRef] [Green Version]
- Szabó, A.; Héja, D.; Szakács, D.; Zboray, K.; Kékesi, K.A.; Radisky, E.S.; Sahin-Tóth, M.; Pál, G. High affinity small protein inhibitors of human chymotrypsin C (CTRC) selected by phage display reveal unusual preference for P4′ acidic residues. J. Biol. Chem. 2011, 286, 22535–22545. [Google Scholar] [CrossRef] [Green Version]
- Szabó, A.; Salameh, M.A.; Ludwig, M.; Radisky, E.S.; Sahin-Tóth, M. Tyrosine sulfation of human trypsin steers S2′ subsite selectivity towards basic amino acids. PLoS ONE 2014, 9, e10206. [Google Scholar] [CrossRef]
- Mahdi, M.; Szojka, Z.; Mótyán, J.A; Tőzsér, J. Inhibition Profiling of Retroviral Protease Inhibitors Using an HIV-2 Modular System. Viruses 2015, 7, 6152–6162. [Google Scholar] [CrossRef] [Green Version]
- Boross, P.; Bagossi, P.; Copeland, T.D.; Oroszlan, S.; Louis, J.M.; Tozser, J. Effect of substrate residues on the P2′ preference of retroviral proteinases. Eur. J. Biochem. 1999, 264, 921–929. [Google Scholar] [CrossRef]
- Ishima, R.; Ghirlando, R.; Tözsér, J.; Gronenborn, A.M.; Torchia, D.A.; Louis, J.M. Folded monomer of HIV-1 protease. J. Biol. Chem. 2001, 276, 49110–49116. [Google Scholar] [CrossRef] [Green Version]
- Mótyán, J.A.; Miczi, M.; Tőzsér, J. Dimer Interface Organization is a Main Determinant of Intermonomeric Interactions and Correlates with Evolutionary Relationships of Retroviral and Retroviral-Like Ddi1 and Ddi2 Proteases. Int. J. Mol. Sci. 2020, 21, 1352. [Google Scholar] [CrossRef] [Green Version]
- Ueda, E.K.M.; Gout, P.W.; Morganti, L. Current and prospective applications of metal ion-protein binding. J. Chromatogr. A 2003, 988, 1–23. [Google Scholar] [CrossRef]
- Voráčková, I.; Suchanová, Š.; Ulbrich, P.; Diehl, W.E.; Ruml, T. Purification of proteins containing zinc finger domains using immobilized metal ion affinity chromatography. Protein Expr. Purif. 2011, 79, 88–95. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wood, R. Atazanavir: Its role in HIV treatment. Expert Rev. Anti Infect. Ther. 2008, 6, 785–796. [Google Scholar] [CrossRef] [Green Version]
- Bagossi, P.; Cheng, Y.S.; Oroszlan, S.; Tozser, J. Comparison of the specificity of homo- and heterodimeric linked HIV-1 and HIV-2 proteinase dimers. Protein Eng. 1998, 11, 439–445. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bagossi, P.; Sperka, T.; Fehér, A.; Kádas, J.; Zahuczky, G.; Miklóssy, G.; Boross, P.; Tözsér, J. Amino acid preferences for a critical substrate binding subsite of retroviral proteases in type 1 cleavage sites. J. Virol. 2005, 79, 4213–4218. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Laco, G.S. HIV-1 protease substrate-groove: Role in substrate recognition and inhibitor resistance. Biochimie 2015, 118, 90–103. [Google Scholar] [CrossRef] [PubMed]
- Laco, G.S. Retroviral proteases: Correlating substrate recognition with both selected and native inhibitor resistance. J. Mol. Biochem. 2017, 6, 45–63. [Google Scholar]
- Kumar, S.; Suguna, K. Crystal structure of the retroviral protease-like domain of a protozoal DNA damage-inducible 1 protein. FEBS Open Bio 2018, 8, 1379–1394. [Google Scholar] [CrossRef] [Green Version]
- Tözsér, J.; Zahuczky, G.; Bagossi, P.; Louis, J.M.; Copeland, T.D.; Oroszlan, S.; Harrison, R.W.; Weber, I.T. Comparison of the substrate specificity of the human T-cell leukemia virus and human immunodeficiency virus proteinases. Eur. J. Biochem. 2000, 267, 6287–6295. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kassay, N.; Mótyán, J.; Matúz, K.; Golda, M.; Tőzsér, J. Biochemical Characterization, Specificity and Inhibition Studies of HTLV-1, HTLV-2, and HTLV-3 Proteases. Life 2021, 11, 127. [Google Scholar] [CrossRef] [PubMed]









| Cleavage Site Name and Sequence | Primer Sequence |
|---|---|
| wt-9res VSQNY*PIVQ | F: 5′-TAAAGTGAGCCAGAACTATCCGATTGTGCAGG-3′ |
| R: 5′-CTAGCCTGCACAATCGGATAGTTCTGGCTCACTTTAAT-3′ | |
| mut-9res VSQLY*PIVQ | F: 5′-TAAAGTGAGCCAGCTGTATCCGATTGTGCAGG-3′ |
| R: 5′-CTAGCCTGCACAATCGGATACAGCTGGCTCACTTTAAT-3′ | |
| wt-24res DTGHSNQVSQNY*PIVQNIQGQMVH | F: 5′-TAAAGATACCGGCCATAGCAACCAGGTGAGCCAGAACTATCCGATTGTGCAGAACATTCAGGGCCAGATGGTGCATG-3′ |
| R: 5′-CTAGCATGCACCATCTGGCCCTGAATGTTCTGCACAATCGGATAGTTCTGGCTCACCTGGTTGCTATGGCCGGTATCTTTAAT–3′ | |
| mut-24res GVGGPGHVSQNY*PIVQSQVTNSAT | F: 5′-TAAAGGCGTGGGCGGCCCGGGCCATGTGAGCCAGAACTATCCGATTGTGCAGAACCAGGTGACCAACAGCGCAACCG-3′ |
| R: 5′-CTAGCGGTTGCGCTGTTGGTCACCTGGTTCTGCACAATCGGATAGTTCTGGCTCACATGGCCCGGGCCGCCCACGCCTTTAAT–3′ |
| Ion | Sample | ka (1/Ms)*103 | kd (1/s)×103 | Requilibrium | Rmax | R2 |
|---|---|---|---|---|---|---|
| nickel | 1 | 0.37 ± 0.12 | 0.42 ± 0.014 | 0.93 | 2.49 | 0.98 |
| 2 | 0.47 ± 0.11 | 0.45 ± 0.012 | 1.08 | 2.54 | 0.98 | |
| cobalt | 1 | 1.08 ± 0.15 | 1.19 ± 0.020 | 0.61 | 1.56 | 0.98 |
| 2 | 2.59 ± 0.15 | 0.69 ± 0.012 | 0.81 | 1.12 | 0.98 | |
| copper | 1 | 0.29 ± 0.14 | 1.85 ± 0.033 | 0.27 | 2.82 | 0.97 |
| 2 | 0.36 ± 0.12 | 1.55 ± 0.060 | 0.45 | 3.26 | 0.97 | |
| zinc | 1 | 1.55 ± 0.12 | 0.81 ± 0.013 | 1.34 | 2.36 | 0.98 |
| 2 | 0.32 ± 0.14 | 0.93 ± 0.021 | 0.68 | 3.53 | 0.97 |
| Substrate | KM (mM)×10−3 | kcat (s−1)×10−3 | kcat/KM (mM−1s−1) |
|---|---|---|---|
| VSQNY*PIVQ 1 | 2.7 ± 0.2 | 50 ± 0.8 | 18.52 ± 1.40 |
| DTGHSNQVSQNY*PIVQNIQGQMVH | 1.68 ± 0.928 | 26.3 ± 1.464 | 15.67 ± 8.71 |
| GVGGPGHVSQNY*PIVQSQVTNSAT | 7.68 ± 0.003 | 40.4 ± 0.005 | 5.26 ± 0.02 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Miczi, M.; Diós, Á.; Bozóki, B.; Tőzsér, J.; Mótyán, J.A. Development of a Bio-Layer Interferometry-Based Protease Assay Using HIV-1 Protease as a Model. Viruses 2021, 13, 1183. https://0-doi-org.brum.beds.ac.uk/10.3390/v13061183
Miczi M, Diós Á, Bozóki B, Tőzsér J, Mótyán JA. Development of a Bio-Layer Interferometry-Based Protease Assay Using HIV-1 Protease as a Model. Viruses. 2021; 13(6):1183. https://0-doi-org.brum.beds.ac.uk/10.3390/v13061183
Chicago/Turabian StyleMiczi, Márió, Ádám Diós, Beáta Bozóki, József Tőzsér, and János András Mótyán. 2021. "Development of a Bio-Layer Interferometry-Based Protease Assay Using HIV-1 Protease as a Model" Viruses 13, no. 6: 1183. https://0-doi-org.brum.beds.ac.uk/10.3390/v13061183