Integrated Proteomics and Transcriptomics Analyses Reveal the Transcriptional Slippage of a Bymovirus P3N-PIPO Gene Expressed from a PVX Vector in Nicotiana benthamiana
Abstract
:1. Introduction
2. Materials and Methods
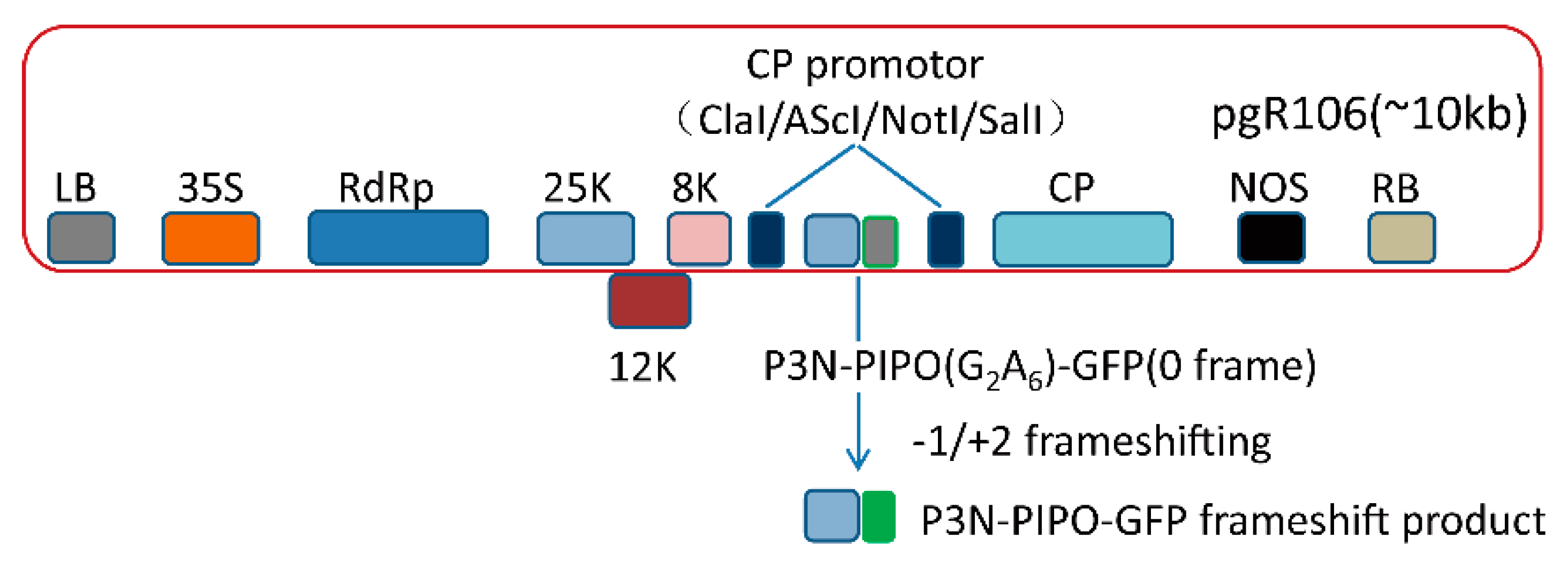
2.1. Construction of PVX-Derived Vectors
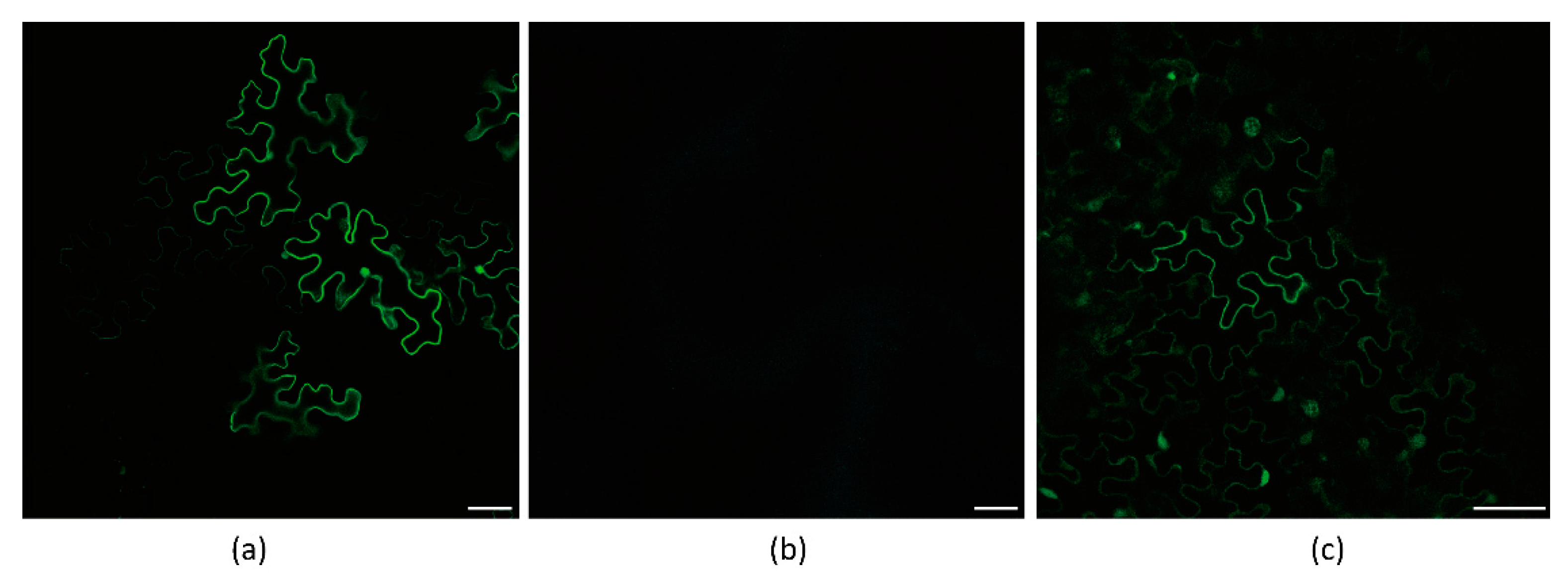
2.2. Growth of Nicotiana Benthamiana Plants, Agrobacterium Infection of Plants, and Confocal Imaging Analysis
2.3. Western Blotting (WB) Analysis
2.4. Protein Purification
2.5. Mass Spectrometric Analysis
2.6. High Throughput Sequencing
3. Results
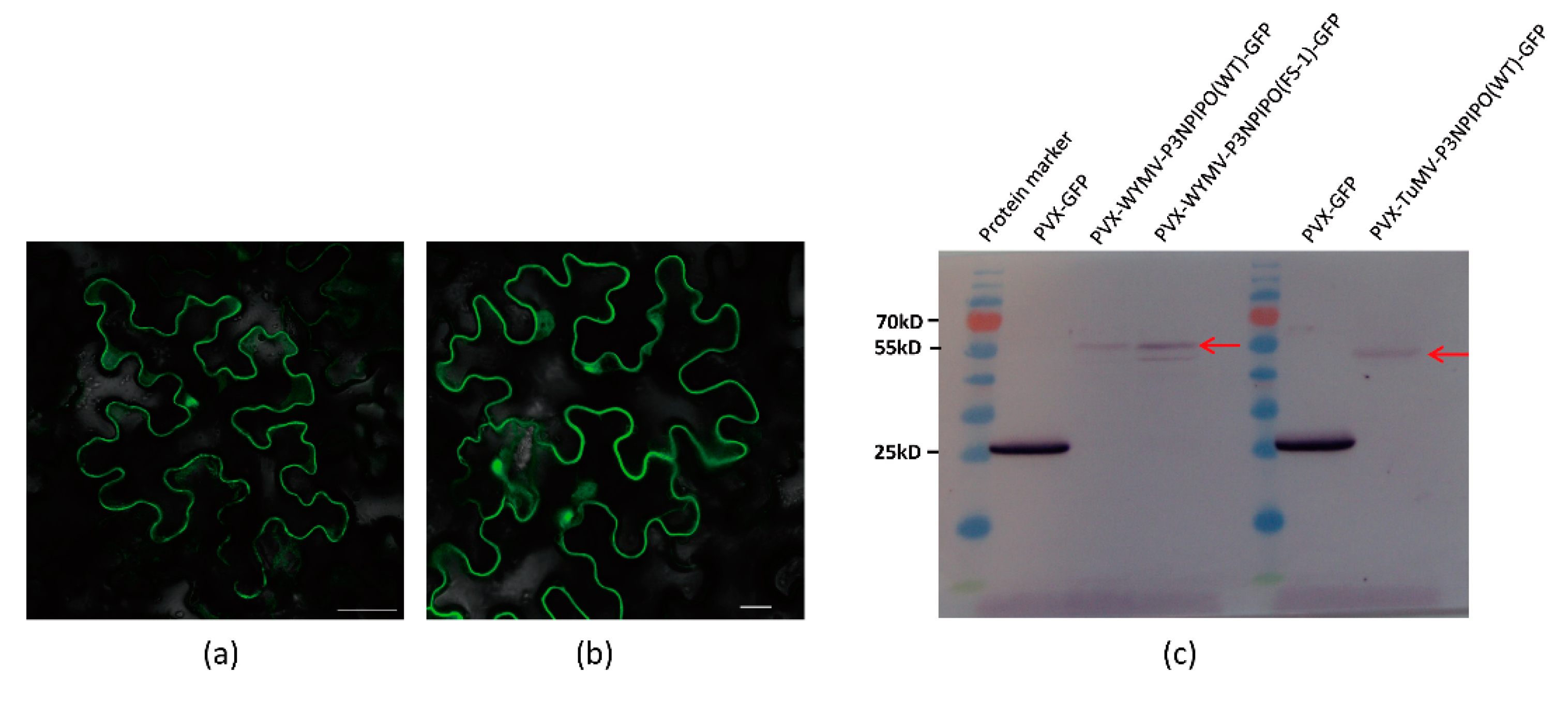
3.1. Frameshift Expression of P3N-PIPO in N. benthamiana by PVX-Based Vector
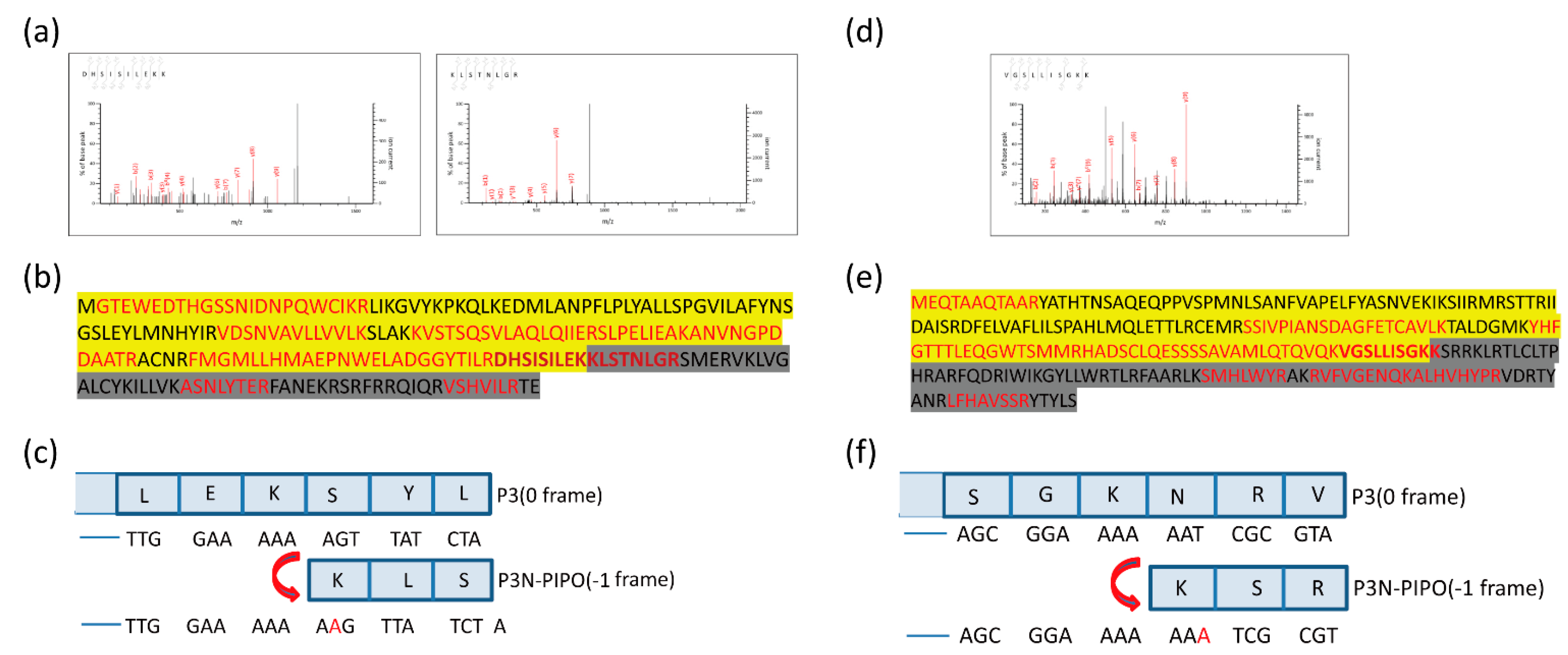
3.2. Mass Spectrometry Revealed That Frameshifting Used in P3N-PIPO(WT)-GFP Expression Took Place at the G2A6 Site
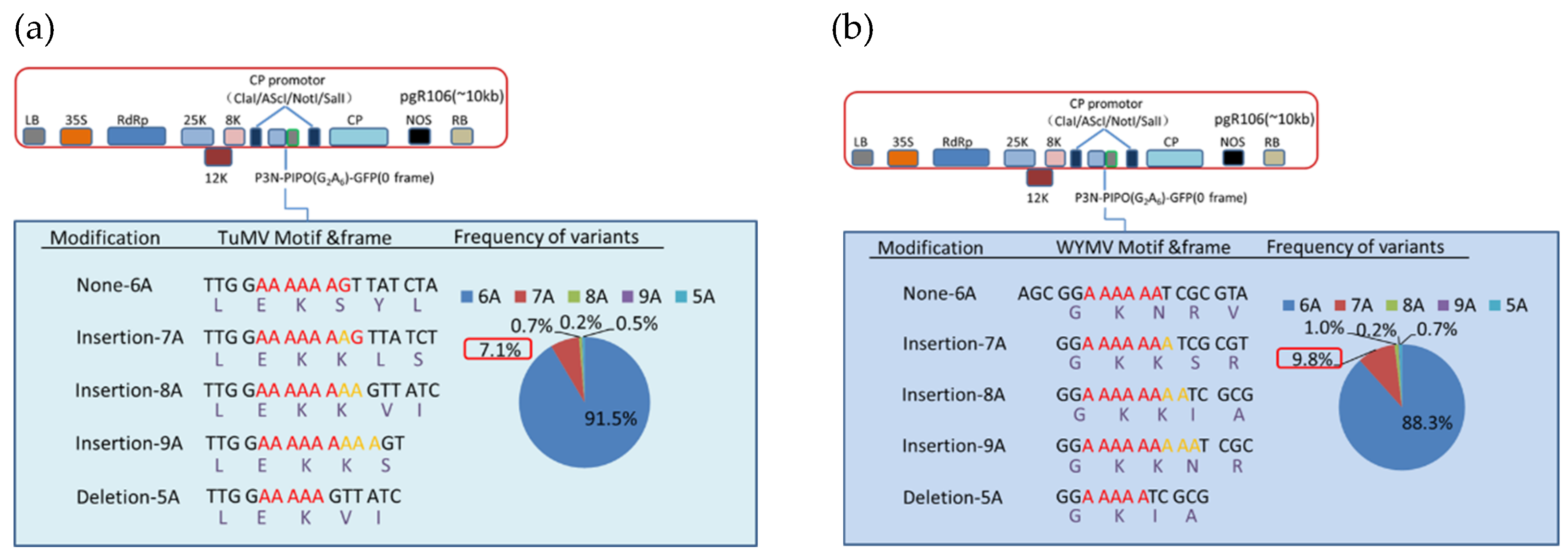
3.3. Deep Sequencing Confirmed the Adenosine Insertion at the G2A6 Site of P3N-PIPO
3.4. Hexa-Adenosine at G2A6 Site Was Necessary for the Frameshift Expression of WYMV P3N-PIPO
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Maia, I.G.; Seron, K.; Haenni, A.-L.; Bernardi, F. Gene Expression from Viral RNA Genomes. Plant Mol. Biol. 1996, 32, 367–391. [Google Scholar] [CrossRef]
- Olspert, A.; Chung, B.Y.; Atkins, J.F.; Carr, J.P.; Firth, A.E. Transcriptional Slippage in the Positive-sense RNA Virus Family Potyviridae. EMBO Rep. 2015, 16, 995–1004. [Google Scholar] [CrossRef] [PubMed]
- Baranov, P.V.; Hammer, A.W.; Zhou, J.; Gesteland, R.F.; Atkins, J.F. Transcriptional Slippage in Bacteria: Distribution in Sequenced Genomes and Utilization in IS Element Gene Expression. Genome Biol. 2005, 6, R25. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wagner, L.A.; Weiss, R.B.; Driscoll, R.; Dunn, D.S.; Gesteland, R.F. Transcriptional Slippage Occurs during Elongation at Runs of Adenine or Thymine in Escherichia Coli. Nucl. Acids Res. 1990, 18, 3529–3535. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, C.; Heath, L.S.; Turnbough, C.L. Regulation of PyrBI Operon Expression in Escherichia Coli by UTP-Sensitive Reiterative RNA Synthesis during Transcriptional Initiation. Genes Dev. 1994, 8, 2904–2912. [Google Scholar] [CrossRef] [Green Version]
- Qi, F.; Turnbough, C.L., Jr. Regulation of codBA Operon Expression In Escherichia Coli by UTP-Dependent Reiterative Transcription and UTP-Sensitive Transcriptional Start Site Switching. J. Mol. Biol. 1995, 254, 552–565. [Google Scholar] [CrossRef]
- Sanchez, A.; Trappier, S.G.; Mahy, B.W.; Peters, C.J.; Nichol, S. The Virion Glycoproteins of Ebola Viruses Are Encoded in Two Reading Frames and Are Expressed through Transcriptional Editing. Proc. Natl. Acad. Sci. USA 1996, 93, 3602–3607. [Google Scholar] [CrossRef] [Green Version]
- Volchkov, V.E.; Becker, S.; Volchkova, V.A.; Ternovoj, V.A.; Kotov, A.N.; Netesov, S.V.; Klenk, H.-D. GP mRNA of Ebola Virus Is Edited by the Ebola Virus Polymerase and by T7 and Vaccinia Virus Polymerases1. Virology 1995, 214, 421–430. [Google Scholar] [CrossRef] [Green Version]
- Volchkova, V.A.; Dolnik, O.; Martinez, M.J.; Reynard, O.; Volchkov, V.E. Genomic RNA Editing and Its Impact on Ebola Virus Adaptation During Serial Passages in Cell Culture and Infection of Guinea Pigs. J. Infect. Dis. 2011, 204, S941–S946. [Google Scholar] [CrossRef]
- Ratinier, M.; Boulant, S.; Combet, C.; Targett-Adams, P.; McLauchlan, J.; Lavergne, J.P. Transcriptional Slippage Prompts Recoding in Alternate Reading Frames in the Hepatitis C Virus (HCV) Core Sequence from Strain HCV-1. J. Gen. Virol. 2008, 89, 1569–1578. [Google Scholar] [CrossRef]
- Hancock, J.M.; Chaleeprom, W.; Chaleeprom, W.; Dale, J.; Gibbs, A. Replication Slippage in the Evolution of Potyviruses. J. Gen. Virol. 1995, 76 Pt 12, 3229–3232. [Google Scholar] [CrossRef]
- Simón-Buela, L.; Osaba, L.; García, J.A.; López-Moya, J.J. Preservation of 5′-End Integrity of a Potyvirus Genomic RNA Is Not Dependent on Template Specificity. Virology 2000, 269, 377–382. [Google Scholar] [CrossRef] [PubMed]
- Chung, B.Y.-W.; Miller, W.A.; Atkins, J.F.; Firth, A.E. An Overlapping Essential Gene in the Potyviridae. Proc. Natl. Acad. Sci. USA 2008, 105, 5897–5902. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vijayapalani, P.; Maeshima, M.; Nagasaki-Takekuchi, N.; Miller, W.A. Interaction of the Trans-Frame Potyvirus Protein P3N-PIPO with Host Protein PCaP1 Facilitates Potyvirus Movement. PLoS Pathog. 2012, 8, e1002639. [Google Scholar] [CrossRef] [PubMed]
- Wei, T.; Zhang, C.; Hong, J.; Xiong, R.; Kasschau, K.D.; Zhou, X.; Carrington, J.C.; Wang, A. Formation of Complexes at Plasmodesmata for Potyvirus Intercellular Movement Is Mediated by the Viral Protein P3N-PIPO. PLoS Pathog. 2010, 6, e1000962. [Google Scholar] [CrossRef] [Green Version]
- Wen, R.-H.; Hajimorad, M.R. Mutational Analysis of the Putative Pipo of Soybean Mosaic Virus Suggests Disruption of PIPO Protein Impedes Movement. Virology 2010, 400, 1–7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Geng, C.; Cong, Q.-Q.; Li, X.-D.; Mou, A.-L.; Gao, R.; Liu, J.-L.; Tian, Y.-P. DEVELOPMENTALLY REGULATED PLASMA MEMBRANE PROTEIN of Nicotiana Benthamiana Contributes to Potyvirus Movement and Transports to Plasmodesmata via the Early Secretory Pathway and the Actomyosin System. Plant Physiol. 2015, 167, 394–410. [Google Scholar] [CrossRef] [Green Version]
- Chai, M.; Wu, X.; Liu, J.; Fang, Y.; Luan, Y.; Cui, X.; Zhou, X.; Wang, A.; Cheng, X. P3N-PIPO Interacts with P3 via the Shared N-Terminal Domain To Recruit Viral Replication Vesicles for Cell-to-Cell Movement. J. Virol. 2020, 94. [Google Scholar] [CrossRef]
- Rodamilans, B.; Valli, A.; Mingot, A.; San León, D.; Baulcombe, D.; López-Moya, J.J.; García, J.A. RNA Polymerase Slippage as a Mechanism for the Production of Frameshift Gene Products in Plant Viruses of the Potyviridae Family. J. Virol. 2015, 89, 6965–6967. [Google Scholar] [CrossRef] [Green Version]
- Chapman, S.; Kavanagh, T.; Baulcombe, D. Potato Virus X as a Vector for Gene Expression in Plants. Plant J. 1992, 2, 549–557. [Google Scholar]
- Dickmeis, C.; Fischer, R.; Commandeur, U. Potato Virus X-Based Expression Vectors Are Stabilized for Long-Term Production of Proteins and Larger Inserts. Biotechnol. J. 2014, 9, 1369–1379. [Google Scholar] [CrossRef] [PubMed]
- Hellens, R.P.; Edwards, E.A.; Leyland, N.R.; Bean, S.; Mullineaux, P.M. PGreen: A Versatile and Flexible Binary Ti Vector for Agrobacterium-Mediated Plant Transformation. Plant Mol. Biol. 2000, 42, 819–832. [Google Scholar] [CrossRef]
- Hayward, A.; Padmanabhan, M.; Dinesh-Kumar, S.P. Virus-Induced Gene Silencing in Nicotiana Benthamiana and Other Plant Species. Methods Mol. Biol. 2011, 678, 55–63. [Google Scholar] [PubMed]
- Granvogl, B.; Plöscher, M.; Eichacker, L.A. Sample Preparation by In-Gel Digestion for Mass Spectrometry-Based Proteomics. Anal. Bioanal. Chem. 2007, 389, 991–1002. [Google Scholar] [CrossRef]
- Thiede, B.; Lamer, S.; Mattow, J.; Siejak, F.; Dimmler, C.; Rudel, T.; Jungblut, P.R. Analysis of Missed Cleavage Sites, Tryptophan Oxidation and N-Terminal Pyroglutamylation after in-Gel Tryptic Digestion. Rapid Commun. Mass Spectrom. 2000, 14, 496–502. [Google Scholar] [CrossRef]
- Valli, A.; García, J.A.; López-Moya, J.J. Potyviruses (Potyviridae). In Encyclopedia of Virology, 4th ed.; Bamford, D.H., Zuckerman, M., Eds.; Academic Press: Oxford, UK, 2021; pp. 631–641. [Google Scholar]
- Untiveros, M.; Olspert, A.; Artola, K.; Firth, A.E.; Kreuze, J.F.; Valkonen, J.P.T. A Novel Sweet Potato Potyvirus Open Reading Frame (ORF) Is Expressed via Polymerase Slippage and Suppresses RNA Silencing. Mol. Plant Pathol. 2016, 17, 1111–1123. [Google Scholar] [CrossRef] [Green Version]
- Mingot, A.; Valli, A.; Rodamilans, B.; San León, D.; Baulcombe, D.C.; García, J.A.; López-Moya, J.J. The P1N-PISPO Trans-Frame Gene of Sweet Potato Feathery Mottle Potyvirus Is Produced during Virus Infection and Functions as an RNA Silencing Suppressor. J. Virol. 2016, 90, 3543–3557. [Google Scholar] [CrossRef] [Green Version]
- White, K.A. The Polymerase Slips and PIPO Exists. EMBO Rep. 2015, 16, 885–886. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yu, C.; Miao, R.; Ye, Z.; MacFarlane, S.; Lu, Y.; Li, J.; Yang, J.; Yan, F.; Dai, L.; Chen, J. Integrated Proteomics and Transcriptomics Analyses Reveal the Transcriptional Slippage of a Bymovirus P3N-PIPO Gene Expressed from a PVX Vector in Nicotiana benthamiana. Viruses 2021, 13, 1247. https://0-doi-org.brum.beds.ac.uk/10.3390/v13071247
Yu C, Miao R, Ye Z, MacFarlane S, Lu Y, Li J, Yang J, Yan F, Dai L, Chen J. Integrated Proteomics and Transcriptomics Analyses Reveal the Transcriptional Slippage of a Bymovirus P3N-PIPO Gene Expressed from a PVX Vector in Nicotiana benthamiana. Viruses. 2021; 13(7):1247. https://0-doi-org.brum.beds.ac.uk/10.3390/v13071247
Chicago/Turabian StyleYu, Chulang, Runpu Miao, Zhuangxin Ye, Stuart MacFarlane, Yuwen Lu, Junmin Li, Jian Yang, Fei Yan, Liangying Dai, and Jianping Chen. 2021. "Integrated Proteomics and Transcriptomics Analyses Reveal the Transcriptional Slippage of a Bymovirus P3N-PIPO Gene Expressed from a PVX Vector in Nicotiana benthamiana" Viruses 13, no. 7: 1247. https://0-doi-org.brum.beds.ac.uk/10.3390/v13071247





