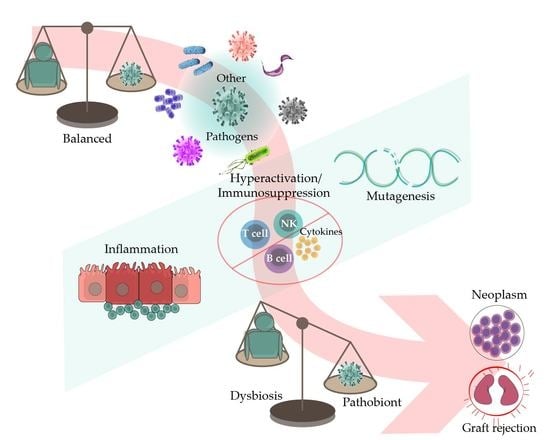
The Role of Coinfections in the EBV–Host Broken Equilibrium
Abstract
:1. Introduction
2. EBV in Immunocompromised Individuals
3. EBV and KSHV Coinfect B Cells in Primary Effusion Lymphoma (PEL) and Germinotropic LPD
4. EBV and Betaherpesviruses in the Transplanted Patient
5. HIV Steady B Cell Stimulation Creates a Lymphomagenesis Permissive Environment
6. Plasmodium falciparum Resembles HIV in Its Capacity to Poly-Clonally Activate B Cells and Trigger GC Continuous Re-Entry
7. EBV Interactions with Oral Bacteria May Facilitate Viral Transmission and Promote Periodontal Diseases
8. HPV Is Also a Tumor Virus That Inhabits Oral and Genital Tissues
9. Helicobacter pylori (Hp), Inflammation, and EBV in the Transformation of Gastric Epithelial Cells
10. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AIDS | Acquired immunodeficiency syndrome |
| ART | antiretroviral therapy |
| BL | Burkitt lymphoma |
| CESC | Cervical squamous cell carcinomas. |
| cHL | Classical Hodgkin lymphoma |
| CMV | Citomegalovirus |
| DLBCL | Diffuse large B cell lymphomas |
| EBV | Epstein–Barr Virus |
| GCR | Germinal center reaction |
| GI | Gastrointestinal |
| GLPD | Germinotropic lymphoproliferative disorders |
| HHV6A | Human herpesvirus 6A |
| HHV6B | Human herpesvirus 6B |
| HHV7 | Human herpesvirus 7 |
| HIV | Human immunodeficiency virus |
| HNSC | Head and neck squamous cell carcinomas |
| HSIL | High-grade squamous intra-epithelial lesion |
| Hp | Helicobacter pylori |
| HPV | Human papillomavirus |
| IARC | International Agency for Research on Cancer |
| ICGC | International Cancer Genome Consortium |
| ICI | Immune checkpoint inhibitors |
| ISH | In situ hybridization |
| KSHV | Kaposi sarcoma virus |
| LPD | Lymphoproliferative disorders |
| LSIL | Low-grade squamous intra-epithelial lesions |
| NPC | Nasopharyngeal carcinoma |
| OR | Odds ratio |
| PCAWG | Pan-Cancer Analysis of Whole Genomes |
| PEL | Primary effusion lymphoma |
| PLWH | People living with HIV |
| PTLD | Post-transplant lymphoproliferative disease |
| TCGA | The Cancer Genome Atlas |
References
- Cruz-Munoz, M.E.; Fuentes-Panana, E.M. Beta and Gamma Human Herpesviruses: Agonistic and Antagonistic Interactions with the Host Immune System. Front. Microbiol. 2017, 8, 2521. [Google Scholar] [CrossRef]
- Thorley-Lawson, D.A. EBV Persistence—Introducing the Virus. Curr. Top. Microbiol. Immunol. 2015, 390, 151–209. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Babcock, G.J.; Decker, L.L.; Volk, M.; Thorley-Lawson, D.A. EBV persistence in memory B cells in vivo. Immunity 1998, 9, 395–404. [Google Scholar] [CrossRef] [Green Version]
- Tangye, S.G.; Palendira, U.; Edwards, E.S. Human immunity against EBV-lessons from the clinic. J. Exp. Med. 2017, 214, 269–283. [Google Scholar] [CrossRef] [PubMed]
- Narkhede, M.; Arora, S.; Ujjani, C. Primary effusion lymphoma: Current perspectives. OncoTargets Ther. 2018, 11, 3747–3754. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jenner, R.G.; Maillard, K.; Cattini, N.; Weiss, R.A.; Boshoff, C.; Wooster, R.; Kellam, P. Kaposi’s sarcoma-associated herpesvirus-infected primary effusion lymphoma has a plasma cell gene expression profile. Proc. Natl. Acad. Sci. USA 2003, 100, 10399–10404. [Google Scholar] [CrossRef] [Green Version]
- Hamoudi, R.; Diss, T.C.; Oksenhendler, E.; Pan, L.; Carbone, A.; Ascoli, V.; Boshoff, C.; Isaacson, P.; Du, M.Q. Distinct cellular origins of primary effusion lymphoma with and without EBV infection. Leuk. Res. 2004, 28, 333–338. [Google Scholar] [CrossRef] [PubMed]
- Roy, D.; Sin, S.H.; Damania, B.; Dittmer, D.P. Tumor suppressor genes FHIT and WWOX are deleted in primary effusion lymphoma (PEL) cell lines. Blood 2011, 118, e32–e39. [Google Scholar] [CrossRef] [Green Version]
- Chadburn, A.; Said, J.; Gratzinger, D.; Chan, J.K.; de Jong, D.; Jaffe, E.S.; Natkunam, Y.; Goodlad, J.R. HHV8/KSHV-Positive Lymphoproliferative Disorders and the Spectrum of Plasmablastic and Plasma Cell Neoplasms: 2015 SH/EAHP Workshop Report-Part 3. Am. J. Clin. Pathol. 2017, 147, 171–187. [Google Scholar] [CrossRef] [Green Version]
- Mesri, E.A.; Cesarman, E.; Arvanitakis, L.; Rafii, S.; Moore, M.A.; Posnett, D.N.; Knowles, D.M.; Asch, A.S. Human herpesvirus-8/Kaposi’s sarcoma-associated herpesvirus is a new transmissible virus that infects B cells. J. Exp. Med. 1996, 183, 2385–2390. [Google Scholar] [CrossRef]
- Faure, A.; Hayes, M.; Sugden, B. How Kaposi’s sarcoma-associated herpesvirus stably transforms peripheral B cells towards lymphomagenesis. Proc. Natl. Acad. Sci. USA 2019, 116, 16519–16528. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bigi, R.; Landis, J.T.; An, H.; Caro-Vegas, C.; Raab-Traub, N.; Dittmer, D.P. Epstein-Barr virus enhances genome maintenance of Kaposi sarcoma-associated herpesvirus. Proc. Natl. Acad. Sci. USA 2018, 115, E11379–E11387. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McHugh, D.; Caduff, N.; Barros, M.H.M.; Ramer, P.C.; Raykova, A.; Murer, A.; Landtwing, V.; Quast, I.; Styles, C.T.; Spohn, M.; et al. Persistent KSHV Infection Increases EBV-Associated Tumor Formation In Vivo via Enhanced EBV Lytic Gene Expression. Cell Host Microbe 2017, 22, 61–73. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Szekely, L.; Chen, F.; Teramoto, N.; Ehlin-Henriksson, B.; Pokrovskaja, K.; Szeles, A.; Manneborg-Sandlund, A.; Lowbeer, M.; Lennette, E.T.; Klein, G. Restricted expression of Epstein-Barr virus (EBV)-encoded, growth transformation-associated antigens in an EBV- and human herpesvirus type 8-carrying body cavity lymphoma line. J. Gen. Virol. 1998, 79, 1445–1452. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krithivas, A.; Young, D.B.; Liao, G.; Greene, D.; Hayward, S.D. Human herpesvirus 8 LANA interacts with proteins of the mSin3 corepressor complex and negatively regulates Epstein-Barr virus gene expression in dually infected PEL cells. J. Virol. 2000, 74, 9637–9645. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miller, G.; Heston, L.; Grogan, E.; Gradoville, L.; Rigsby, M.; Sun, R.; Shedd, D.; Kushnaryov, V.M.; Grossberg, S.; Chang, Y. Selective switch between latency and lytic replication of Kaposi’s sarcoma herpesvirus and Epstein-Barr virus in dually infected body cavity lymphoma cells. J. Virol. 1997, 71, 314–324. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Groves, A.K.; Cotter, M.A.; Subramanian, C.; Robertson, E.S. The latency-associated nuclear antigen encoded by Kaposi’s sarcoma-associated herpesvirus activates two major essential Epstein-Barr virus latent promoters. J. Virol. 2001, 75, 9446–9457. [Google Scholar] [CrossRef] [Green Version]
- Xu, D.; Coleman, T.; Zhang, J.; Fagot, A.; Kotalik, C.; Zhao, L.; Trivedi, P.; Jones, C.; Zhang, L. Epstein-Barr virus inhibits Kaposi’s sarcoma-associated herpesvirus lytic replication in primary effusion lymphomas. J. Virol. 2007, 81, 6068–6078. [Google Scholar] [CrossRef] [Green Version]
- Spadavecchia, S.; Gonzalez-Lopez, O.; Carroll, K.D.; Palmeri, D.; Lukac, D.M. Convergence of Kaposi’s sarcoma-associated herpesvirus reactivation with Epstein-Barr virus latency and cellular growth mediated by the notch signaling pathway in coinfected cells. J. Virol. 2010, 84, 10488–10500. [Google Scholar] [CrossRef] [Green Version]
- Jiang, Y.; Xu, D.; Zhao, Y.; Zhang, L. Mutual inhibition between Kaposi’s sarcoma-associated herpesvirus and Epstein-Barr virus lytic replication initiators in dually-infected primary effusion lymphoma. PLoS ONE 2008, 3, e1569. [Google Scholar] [CrossRef]
- Horenstein, M.G.; Nador, R.G.; Chadburn, A.; Hyjek, E.M.; Inghirami, G.; Knowles, D.M.; Cesarman, E. Epstein-Barr virus latent gene expression in primary effusion lymphomas containing Kaposi’s sarcoma-associated herpesvirus/human herpesvirus-8. Blood 1997, 90, 1186–1191. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Callahan, J.; Pai, S.; Cotter, M.; Robertson, E.S. Distinct patterns of viral antigen expression in Epstein-Barr virus and Kaposi’s sarcoma-associated herpesvirus coinfected body-cavity-based lymphoma cell lines: Potential switches in latent gene expression due to coinfection. Virology 1999, 262, 18–30. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choi, U.Y.; Park, A.; Jung, J.U. Double the Trouble When Herpesviruses Join Hands. Cell Host Microbe 2017, 22, 5–7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Riva, G.; Lagreca, I.; Mattiolo, A.; Belletti, D.; Lignitto, L.; Barozzi, P.; Ruozi, B.; Vallerini, D.; Quadrelli, C.; Corradini, G.; et al. Antineoplastic effects of liposomal short interfering RNA treatment targeting BLIMP1/PRDM1 in primary effusion lymphoma. Haematologica 2015, 100, e467–e470. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reusch, J.A.; Nawandar, D.M.; Wright, K.L.; Kenney, S.C.; Mertz, J.E. Cellular differentiation regulator BLIMP1 induces Epstein-Barr virus lytic reactivation in epithelial and B cells by activating transcription from both the R and Z promoters. J. Virol. 2015, 89, 1731–1743. [Google Scholar] [CrossRef] [Green Version]
- Shapiro-Shelef, M.; Lin, K.I.; McHeyzer-Williams, L.J.; Liao, J.; McHeyzer-Williams, M.G.; Calame, K. Blimp-1 is required for the formation of immunoglobulin secreting plasma cells and pre-plasma memory B cells. Immunity 2003, 19, 607–620. [Google Scholar] [CrossRef] [Green Version]
- Trivedi, P.; Takazawa, K.; Zompetta, C.; Cuomo, L.; Anastasiadou, E.; Carbone, A.; Uccini, S.; Belardelli, F.; Takada, K.; Frati, L.; et al. Infection of HHV-8+ primary effusion lymphoma cells with a recombinant Epstein-Barr virus leads to restricted EBV latency, altered phenotype, and increased tumorigenicity without affecting TCL1 expression. Blood 2004, 103, 313–316. [Google Scholar] [CrossRef] [Green Version]
- Mack, A.A.; Sugden, B. EBV is necessary for proliferation of dually infected primary effusion lymphoma cells. Cancer Res. 2008, 68, 6963–6968. [Google Scholar] [CrossRef] [Green Version]
- Dittmer, D.P.; Damania, B. Kaposi sarcoma-associated herpesvirus: Immunobiology, oncogenesis, and therapy. J. Clin. Investig. 2016, 126, 3165–3175. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shin, Y.C.; Nakamura, H.; Liang, X.; Feng, P.; Chang, H.; Kowalik, T.F.; Jung, J.U. Inhibition of the ATM/p53 signal transduction pathway by Kaposi’s sarcoma-associated herpesvirus interferon regulatory factor 1. J. Virol. 2006, 80, 2257–2266. [Google Scholar] [CrossRef] [Green Version]
- Seo, T.; Park, J.; Choe, J. Kaposi’s sarcoma-associated herpesvirus viral IFN regulatory factor 1 inhibits transforming growth factor-beta signaling. Cancer Res. 2005, 65, 1738–1747. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, H.J.; Kim, Y.M.; Lim, S.; Nam, Y.K.; Jeong, J.; Kim, H.J.; Lee, K.J. Ubiquitin C-terminal hydrolase-L1 is a key regulator of tumor cell invasion and metastasis. Oncogene 2009, 28, 117–127. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Otsuki, T.; Yata, K.; Takata-Tomokuni, A.; Hyodoh, F.; Miura, Y.; Sakaguchi, H.; Hatayama, T.; Hatada, S.; Tsujioka, T.; Sato, Y.; et al. Expression of protein gene product 9.5 (PGP9.5)/ubiquitin-C-terminal hydrolase 1 (UCHL-1) in human myeloma cells. Br. J. Haematol. 2004, 127, 292–298. [Google Scholar] [CrossRef]
- Ovaa, H.; Kessler, B.M.; Rolen, U.; Galardy, P.J.; Ploegh, H.L.; Masucci, M.G. Activity-based ubiquitin-specific protease (USP) profiling of virus-infected and malignant human cells. Proc. Natl. Acad. Sci. USA 2004, 101, 2253–2258. [Google Scholar] [CrossRef] [Green Version]
- Bentz, G.L.; Bheda-Malge, A.; Wang, L.; Shackelford, J.; Damania, B.; Pagano, J.S. KSHV LANA and EBV LMP1 induce the expression of UCH-L1 following viral transformation. Virology 2014, 448, 293–302. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gruffat, H.; Manet, E. EBV/KSHV co-infection: An effective partnership. Med. Sci. 2018, 34, 79–82. [Google Scholar] [CrossRef]
- Ma, S.D.; Hegde, S.; Young, K.H.; Sullivan, R.; Rajesh, D.; Zhou, Y.; Jankowska-Gan, E.; Burlingham, W.J.; Sun, X.; Gulley, M.L.; et al. A new model of Epstein-Barr virus infection reveals an important role for early lytic viral protein expression in the development of lymphomas. J. Virol. 2011, 85, 165–177. [Google Scholar] [CrossRef] [Green Version]
- Gloghini, A.; Volpi, C.C.; Gualeni, A.V.; Dolcetti, R.; Bongarzone, I.; De Paoli, P.; Carbone, A. Multiple viral infections in primary effusion lymphoma: A model of viral cooperation in lymphomagenesis. Expert Rev. Hematol. 2017, 10, 505–514. [Google Scholar] [CrossRef]
- Schulz, T.F.; Cesarman, E. Kaposi Sarcoma-associated Herpesvirus: Mechanisms of oncogenesis. Curr. Opin. Virol. 2015, 14, 116–128. [Google Scholar] [CrossRef]
- Pantanowitz, L.; Carbone, A.; Dolcetti, R. Microenvironment and HIV-related lymphomagenesis. Semin. Cancer Biol. 2015, 34, 52–57. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, M.; Osborne, J.; Bestetti, G.; Chang, Y.; Moore, P.S. Viral IL-6-induced cell proliferation and immune evasion of interferon activity. Science 2002, 298, 1432–1435. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jones, K.D.; Aoki, Y.; Chang, Y.; Moore, P.S.; Yarchoan, R.; Tosato, G. Involvement of interleukin-10 (IL-10) and viral IL-6 in the spontaneous growth of Kaposi’s sarcoma herpesvirus-associated infected primary effusion lymphoma cells. Blood 1999, 94, 2871–2879. [Google Scholar] [CrossRef] [PubMed]
- Punj, V.; Matta, H.; Schamus, S.; Yang, T.; Chang, Y.; Chaudhary, P.M. Induction of CCL20 production by Kaposi sarcoma-associated herpesvirus: Role of viral FLICE inhibitory protein K13-induced NF-kappaB activation. Blood 2009, 113, 5660–5668. [Google Scholar] [CrossRef] [Green Version]
- Cinatl, J., Jr.; Vogel, J.U.; Kotchetkov, R.; Wilhelm Doerr, H. Oncomodulatory signals by regulatory proteins encoded by human cytomegalovirus: A novel role for viral infection in tumor progression. FEMS Microbiol. Rev. 2004, 28, 59–77. [Google Scholar] [CrossRef] [Green Version]
- Flamand, L.; Gosselin, J.; Stefanescu, I.; Ablashi, D.; Menezes, J. Immunosuppressive effect of human herpesvirus 6 on T-cell functions: Suppression of interleukin-2 synthesis and cell proliferation. Blood 1995, 85, 1263–1271. [Google Scholar] [CrossRef]
- Ogata, M. Human herpesvirus 6 in hematological malignancies. J. Clin. Exp. Hematop. 2009, 49, 57–67. [Google Scholar] [CrossRef] [Green Version]
- Jenkins, F.J.; Rowe, D.T.; Rinaldo, C.R., Jr. Herpesvirus infections in organ transplant recipients. Clin. Diagn. Lab. Immunol. 2003, 10, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Cohen, J.I. Chapter 142—Human Herpesvirus Types 6 and 7 (Exanthem Subitum). In Mandell, Douglas, and Bennett’s Principles and Practice of Infectious Diseases, 8th ed.; Bennett, J.E., Dolin, R., Blaser, M.J., Eds.; W.B. Saunders: Philadelphia, PA, USA, 2015; pp. 1772–1776.e1. [Google Scholar]
- Mendez, J.C.; Dockrell, D.H.; Espy, M.J.; Smith, T.F.; Wilson, J.A.; Harmsen, W.S.; Ilstrup, D.; Paya, C.V. Human beta-herpesvirus interactions in solid organ transplant recipients. J. Infect. Dis. 2001, 183, 179–184. [Google Scholar] [CrossRef] [PubMed]
- Razonable, R.R.; Humar, A.; Practice, A.S.T.I.D.C.o. Cytomegalovirus in solid organ transplantation. Am. J. Transplant. 2013, 13, 93–106. [Google Scholar] [CrossRef] [PubMed]
- Anderson-Smits, C.; Baker, E.R.; Hirji, I. Coinfection rates and clinical outcome data for cytomegalovirus and Epstein-Barr virus in post-transplant patients: A systematic review of the literature. Transpl. Infect. Dis. 2020, 22, e13396. [Google Scholar] [CrossRef]
- Indolfi, G.; Heaton, N.; Smith, M.; Mieli-Vergani, G.; Zuckerman, M. Effect of early EBV and/or CMV viremia on graft function and acute cellular rejection in pediatric liver transplantation. Clin. Transplant. 2012, 26, E55–E61. [Google Scholar] [CrossRef]
- Ono, Y.; Ito, Y.; Kaneko, K.; Shibata-Watanabe, Y.; Tainaka, T.; Sumida, W.; Nakamura, T.; Kamei, H.; Kiuchi, T.; Ando, H.; et al. Simultaneous monitoring by real-time polymerase chain reaction of epstein-barr virus, human cytomegalovirus, and human herpesvirus-6 in juvenile and adult liver transplant recipients. Transplant. Proc. 2008, 40, 3578–3582. [Google Scholar] [CrossRef]
- Garcia-Cadenas, I.; Castillo, N.; Martino, R.; Barba, P.; Esquirol, A.; Novelli, S.; Orti, G.; Garrido, A.; Saavedra, S.; Moreno, C.; et al. Impact of Epstein Barr virus-related complications after high-risk allo-SCT in the era of pre-emptive rituximab. Bone Marrow Transplant. 2015, 50, 579–584. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barani, R.; Ravi, Y.; Seshan, V.; Reju, S.; Soundararajan, P.; Palani, G.; Srikanth, P. Epstein-Barr Virus DNAemia and co-occurrence with cytomegalovirus DNAemia in postrenal transplant recipients from a tertiary care center. Indian J. Transplant. 2018, 12, 95–102. [Google Scholar] [CrossRef]
- Shivanesan, P.; Minz, M.; Minz, R.W.; Kumar, Y.; Sharma, A.; Kanwar, D.B.; Singh, S.; Kohli, H.S.; Anand, S.; Nada, R. Utility of quantitative real time PCR in detection and monitoring of viral infections in post renal transplant recipients. Indian J. Transplant. 2016, 10, 9–14. [Google Scholar] [CrossRef]
- Sanchez-Ponce, Y.; Varela-Fascinetto, G.; Romo-Vazquez, J.C.; Lopez-Martinez, B.; Sanchez-Huerta, J.L.; Parra-Ortega, I.; Fuentes-Panana, E.M.; Morales-Sanchez, A. Simultaneous Detection of Beta and Gamma Human Herpesviruses by Multiplex qPCR Reveals Simple Infection and Coinfection Episodes Increasing Risk for Graft Rejection in Solid Organ Transplantation. Viruses 2018, 10, 730. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Al-Mansour, Z.; Nelson, B.P.; Evens, A.M. Post-transplant lymphoproliferative disease (PTLD): Risk factors, diagnosis, and current treatment strategies. Curr. Hematol. Malig. Rep. 2013, 8, 173–183. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lam, J.K.P.; Azzi, T.; Hui, K.F.; Wong, A.M.G.; McHugh, D.; Caduff, N.; Chan, K.H.; Munz, C.; Chiang, A.K.S. Co-infection of Cytomegalovirus and Epstein-Barr Virus Diminishes the Frequency of CD56(dim)NKG2A(+)KIR(-) NK Cells and Contributes to Suboptimal Control of EBV in Immunosuppressed Children With Post-transplant Lymphoproliferative Disorder. Front. Immunol. 2020, 11, 1231. [Google Scholar] [CrossRef]
- Zallio, F.; Primon, V.; Tamiazzo, S.; Pini, M.; Baraldi, A.; Corsetti, M.T.; Gotta, F.; Bertassello, C.; Salvi, F.; Rocchetti, A.; et al. Epstein-Barr virus reactivation in allogeneic stem cell transplantation is highly related to cytomegalovirus reactivation. Clin. Transplant. 2013, 27, E491–E497. [Google Scholar] [CrossRef]
- Allen, U.D.; Preiksaitis, J.K.; Practice, A.S.T.I.D.C.o. Post-transplant lymphoproliferative disorders, Epstein-Barr virus infection, and disease in solid organ transplantation: Guidelines from the American Society of Transplantation Infectious Diseases Community of Practice. Clin. Transplant. 2019, 33, e13652. [Google Scholar] [CrossRef] [PubMed]
- Kimura, H.; Kwong, Y.L. EBV Viral Loads in Diagnosis, Monitoring, and Response Assessment. Front. Oncol. 2019, 9, 62. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kimura, H.; Ito, Y.; Suzuki, R.; Nishiyama, Y. Measuring Epstein-Barr virus (EBV) load: The significance and application for each EBV-associated disease. Rev. Med. Virol. 2008, 18, 305–319. [Google Scholar] [CrossRef]
- Wadowsky, R.M.; Laus, S.; Green, M.; Webber, S.A.; Rowe, D. Measurement of Epstein-Barr virus DNA loads in whole blood and plasma by TaqMan PCR and in peripheral blood lymphocytes by competitive PCR. J. Clin. Microbiol. 2003, 41, 5245–5249. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.; Yang, K.; Wei, C.; Huang, Y.; Zhao, D. Coinfection with EBV/CMV and other respiratory agents in children with suspected infectious mononucleosis. Virol. J. 2010, 7, 247. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ito, Y.; Shibata-Watanabe, Y.; Kawada, J.; Maruyama, K.; Yagasaki, H.; Kojima, S.; Kimura, H. Cytomegalovirus and Epstein-Barr virus coinfection in three toddlers with prolonged illnesses. J. Med. Virol. 2009, 81, 1399–1402. [Google Scholar] [CrossRef]
- Olson, D.; Huntington, M.K. Co-infection with cytomegalovirus and Epstein-Barr virus in mononucleosis: Case report and review of literature. S D Med. 2009, 62, 349, 351–353. [Google Scholar]
- Abate, F.; Ambrosio, M.R.; Mundo, L.; Laginestra, M.A.; Fuligni, F.; Rossi, M.; Zairis, S.; Gazaneo, S.; De Falco, G.; Lazzi, S.; et al. Distinct Viral and Mutational Spectrum of Endemic Burkitt Lymphoma. PLoS Pathog. 2015, 11, e1005158. [Google Scholar] [CrossRef] [Green Version]
- Saghafian-Hedengren, S.; Sundstrom, Y.; Sohlberg, E.; Nilsson, C.; Linde, A.; Troye-Blomberg, M.; Berg, L.; Sverremark-Ekstrom, E. Herpesvirus seropositivity in childhood associates with decreased monocyte-induced NK cell IFN-gamma production. J. Immunol. 2009, 182, 2511–2517. [Google Scholar] [CrossRef]
- Muller-Durovic, B.; Grahlert, J.; Devine, O.P.; Akbar, A.N.; Hess, C. CD56-negative NK cells with impaired effector function expand in CMV and EBV co-infected healthy donors with age. Aging 2019, 11, 724–740. [Google Scholar] [CrossRef]
- Saghafian-Hedengren, S.; Sohlberg, E.; Theorell, J.; Carvalho-Queiroz, C.; Nagy, N.; Persson, J.O.; Nilsson, C.; Bryceson, Y.T.; Sverremark-Ekstrom, E. Epstein-Barr virus coinfection in children boosts cytomegalovirus-induced differentiation of natural killer cells. J. Virol. 2013, 87, 13446–13455. [Google Scholar] [CrossRef] [Green Version]
- Sohlberg, E.; Saghafian-Hedengren, S.; Rasul, E.; Marchini, G.; Nilsson, C.; Klein, E.; Nagy, N.; Sverremark-Ekstrom, E. Cytomegalovirus-seropositive children show inhibition of in vitro EBV infection that is associated with CD8+CD57+ T cell enrichment and IFN-gamma. J. Immunol. 2013, 191, 5669–5676. [Google Scholar] [CrossRef] [Green Version]
- Gosselin, J.; Flamand, L.; D’Addario, M.; Hiscott, J.; Stefanescu, I.; Ablashi, D.V.; Gallo, R.C.; Menezes, J. Modulatory effects of Epstein-Barr, herpes simplex, and human herpes-6 viral infections and coinfections on cytokine synthesis. A comparative study. J. Immunol. 1992, 149, 181–187. [Google Scholar]
- Flamand, L.; Stefanescu, I.; Ablashi, D.V.; Menezes, J. Activation of the Epstein-Barr virus replicative cycle by human herpesvirus 6. J. Virol. 1993, 67, 6768–6777. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Simard, E.P.; Engels, E.A. Cancer as a cause of death among people with AIDS in the United States. Clin. Infect. Dis. 2010, 51, 957–962. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shindiapina, P.; Ahmed, E.H.; Mozhenkova, A.; Abebe, T.; Baiocchi, R.A. Immunology of EBV-Related Lymphoproliferative Disease in HIV-Positive Individuals. Front. Oncol. 2020, 10, 1723. [Google Scholar] [CrossRef]
- Simard, E.P.; Pfeiffer, R.M.; Engels, E.A. Cumulative incidence of cancer among individuals with acquired immunodeficiency syndrome in the United States. Cancer 2011, 117, 1089–1096. [Google Scholar] [CrossRef]
- Carbone, A.; Gloghini, A.; Larocca, L.M.; Antinori, A.; Falini, B.; Tirelli, U.; Dalla-Favera, R.; Gaidano, G. Human immunodeficiency virus-associated Hodgkin’s disease derives from post-germinal center B cells. Blood 1999, 93, 2319–2326. [Google Scholar] [PubMed]
- Gibson, T.M.; Morton, L.M.; Shiels, M.S.; Clarke, C.A.; Engels, E.A. Risk of non-Hodgkin lymphoma subtypes in HIV-infected people during the HAART era: A population-based study. AIDS 2014, 28, 2313–2318. [Google Scholar] [CrossRef] [Green Version]
- Mbulaiteye, S.M.; Pullarkat, S.T.; Nathwani, B.N.; Weiss, L.M.; Rao, N.; Emmanuel, B.; Lynch, C.F.; Hernandez, B.; Neppalli, V.; Hawes, D.; et al. Epstein-Barr virus patterns in US Burkitt lymphoma tumors from the SEER residual tissue repository during 1979-2009. APMIS 2014, 122, 5–15. [Google Scholar] [CrossRef] [Green Version]
- Kersten, M.J.; Van Gorp, J.; Pals, S.T.; Boon, F.; Van Oers, M.H. Expression of Epstein-Barr virus latent genes and adhesion molecules in AIDS-related non-Hodgkin’s lymphomas: Correlation with histology and CD4-cell number. Leuk. Lymphoma 1998, 30, 515–524. [Google Scholar] [CrossRef]
- Linke-Serinsoz, E.; Fend, F.; Quintanilla-Martinez, L. Human immunodeficiency virus (HIV) and Epstein-Barr virus (EBV) related lymphomas, pathology view point. Semin. Diagn. Pathol. 2017, 34, 352–363. [Google Scholar] [CrossRef]
- Carbone, A.; Gloghini, A.; Caruso, A.; De Paoli, P.; Dolcetti, R. The impact of EBV and HIV infection on the microenvironmental niche underlying Hodgkin lymphoma pathogenesis. Int. J. Cancer 2017, 140, 1233–1245. [Google Scholar] [CrossRef]
- Kelly, G.L.; Stylianou, J.; Rasaiyaah, J.; Wei, W.; Thomas, W.; Croom-Carter, D.; Kohler, C.; Spang, R.; Woodman, C.; Kellam, P.; et al. Different patterns of Epstein-Barr virus latency in endemic Burkitt lymphoma (BL) lead to distinct variants within the BL-associated gene expression signature. J. Virol. 2013, 87, 2882–2894. [Google Scholar] [CrossRef] [Green Version]
- Dolcetti, R.; Gloghini, A.; Caruso, A.; Carbone, A. A lymphomagenic role for HIV beyond immune suppression? Blood 2016, 127, 1403–1409. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vrzalikova, K.; Woodman, C.B.; Murray, P.G. BLIMP1alpha, the master regulator of plasma cell differentiation is a tumor supressor gene in B cell lymphomas. Biomed. Pap. Med. Fac. Univ. Palacky Olomouc Czech Repub. 2012, 156, 1–6. [Google Scholar] [CrossRef]
- Vrzalikova, K.; Vockerodt, M.; Leonard, S.; Bell, A.; Wei, W.; Schrader, A.; Wright, K.L.; Kube, D.; Rowe, M.; Woodman, C.B.; et al. Down-regulation of BLIMP1alpha by the EBV oncogene, LMP-1, disrupts the plasma cell differentiation program and prevents viral replication in B cells: Implications for the pathogenesis of EBV-associated B-cell lymphomas. Blood 2011, 117, 5907–5917. [Google Scholar] [CrossRef] [PubMed]
- Malaspina, A.; Moir, S.; Kottilil, S.; Hallahan, C.W.; Ehler, L.A.; Liu, S.; Planta, M.A.; Chun, T.W.; Fauci, A.S. Deleterious effect of HIV-1 plasma viremia on B cell costimulatory function. J. Immunol. 2003, 170, 5965–5972. [Google Scholar] [CrossRef] [Green Version]
- Nagase, H.; Agematsu, K.; Kitano, K.; Takamoto, M.; Okubo, Y.; Komiyama, A.; Sugane, K. Mechanism of hypergammaglobulinemia by HIV infection: Circulating memory B-cell reduction with plasmacytosis. Clin. Immunol. 2001, 100, 250–259. [Google Scholar] [CrossRef] [PubMed]
- Curreli, S.; Krishnan, S.; Reitz, M.; Lunardi-Iskandar, Y.; Lafferty, M.K.; Garzino-Demo, A.; Zella, D.; Gallo, R.C.; Bryant, J. B cell lymphoma in HIV transgenic mice. Retrovirology 2013, 10, 92. [Google Scholar] [CrossRef] [Green Version]
- Kundu, R.K.; Sangiorgi, F.; Wu, L.Y.; Pattengale, P.K.; Hinton, D.R.; Gill, P.S.; Maxson, R. Expression of the human immunodeficiency virus-Tat gene in lymphoid tissues of transgenic mice is associated with B-cell lymphoma. Blood 1999, 94, 275–282. [Google Scholar] [CrossRef]
- Popovic, M.; Tenner-Racz, K.; Pelser, C.; Stellbrink, H.J.; van Lunzen, J.; Lewis, G.; Kalyanaraman, V.S.; Gallo, R.C.; Racz, P. Persistence of HIV-1 structural proteins and glycoproteins in lymph nodes of patients under highly active antiretroviral therapy. Proc. Natl. Acad. Sci. USA 2005, 102, 14807–14812. [Google Scholar] [CrossRef] [Green Version]
- Lazzi, S.; Bellan, C.; De Falco, G.; Cinti, C.; Ferrari, F.; Nyongo, A.; Claudio, P.P.; Tosi, G.M.; Vatti, R.; Gloghini, A.; et al. Expression of RB2/p130 tumor-suppressor gene in AIDS-related non-Hodgkin’s lymphomas: Implications for disease pathogenesis. Hum. Pathol. 2002, 33, 723–731. [Google Scholar] [CrossRef] [Green Version]
- Giagulli, C.; Marsico, S.; Magiera, A.K.; Bruno, R.; Caccuri, F.; Barone, I.; Fiorentini, S.; Ando, S.; Caruso, A. Opposite effects of HIV-1 p17 variants on PTEN activation and cell growth in B cells. PLoS ONE 2011, 6, e17831. [Google Scholar] [CrossRef] [PubMed]
- Caccuri, F.; Giagulli, C.; Reichelt, J.; Martorelli, D.; Marsico, S.; Bugatti, A.; Barone, I.; Rusnati, M.; Guzman, C.A.; Dolcetti, R.; et al. Simian immunodeficiency virus and human immunodeficiency virus type 1 matrix proteins specify different capabilities to modulate B cell growth. J. Virol. 2014, 88, 5706–5717. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martin, G.; Tremblay, M.J. HLA-DR, ICAM-1, CD40, CD40L, and CD86 are incorporated to a similar degree into clinical human immunodeficiency virus type 1 variants expanded in natural reservoirs such as peripheral blood mononuclear cells and human lymphoid tissue cultured ex vivo. Clin. Immunol. 2004, 111, 275–285. [Google Scholar] [CrossRef]
- Klein, U.; Dalla-Favera, R. Germinal centres: Role in B-cell physiology and malignancy. Nat. Rev. Immunol. 2008, 8, 22–33. [Google Scholar] [CrossRef] [PubMed]
- Martin, G.; Roy, J.; Barat, C.; Ouellet, M.; Gilbert, C.; Tremblay, M.J. Human immunodeficiency virus type 1-associated CD40 ligand transactivates B lymphocytes and promotes infection of CD4+ T cells. J. Virol. 2007, 81, 5872–5881. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Imbeault, M.; Ouellet, M.; Giguere, K.; Bertin, J.; Belanger, D.; Martin, G.; Tremblay, M.J. Acquisition of host-derived CD40L by HIV-1 in vivo and its functional consequences in the B-cell compartment. J. Virol. 2011, 85, 2189–2200. [Google Scholar] [CrossRef] [Green Version]
- Fish, K.; Comoglio, F.; Shaffer, A.L., 3rd; Ji, Y.; Pan, K.T.; Scheich, S.; Oellerich, A.; Doebele, C.; Ikeda, M.; Schaller, S.J.; et al. Rewiring of B cell receptor signaling by Epstein-Barr virus LMP2A. Proc. Natl. Acad. Sci. USA 2020, 117, 26318–26327. [Google Scholar] [CrossRef]
- Mancao, C.; Altmann, M.; Jungnickel, B.; Hammerschmidt, W. Rescue of “crippled” germinal center B cells from apoptosis by Epstein-Barr virus. Blood 2005, 106, 4339–4344. [Google Scholar] [CrossRef] [PubMed]
- Martorelli, D.; Muraro, E.; Mastorci, K.; Dal Col, J.; Fae, D.A.; Furlan, C.; Giagulli, C.; Caccuri, F.; Rusnati, M.; Fiorentini, S.; et al. A natural HIV p17 protein variant up-regulates the LMP-1 EBV oncoprotein and promotes the growth of EBV-infected B-lymphocytes: Implications for EBV-driven lymphomagenesis in the HIV setting. Int. J. Cancer 2015, 137, 1374–1385. [Google Scholar] [CrossRef]
- Lam, N.; Sugden, B. CD40 and its viral mimic, LMP1: Similar means to different ends. Cell Signal. 2003, 15, 9–16. [Google Scholar] [CrossRef]
- Wada, N.I.; Jacobson, L.P.; Margolick, J.B.; Breen, E.C.; Macatangay, B.; Penugonda, S.; Martinez-Maza, O.; Bream, J.H. The effect of HAART-induced HIV suppression on circulating markers of inflammation and immune activation. AIDS 2015, 29, 463–471. [Google Scholar] [CrossRef] [Green Version]
- Harker, J.A.; Lewis, G.M.; Mack, L.; Zuniga, E.I. Late interleukin-6 escalates T follicular helper cell responses and controls a chronic viral infection. Science 2011, 334, 825–829. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rousset, F.; Garcia, E.; Defrance, T.; Peronne, C.; Vezzio, N.; Hsu, D.H.; Kastelein, R.; Moore, K.W.; Banchereau, J. Interleukin 10 is a potent growth and differentiation factor for activated human B lymphocytes. Proc. Natl. Acad. Sci. USA 1992, 89, 1890–1893. [Google Scholar] [CrossRef] [Green Version]
- Bishop, G.A.; Hostager, B.S. The CD40-CD154 interaction in B cell-T cell liaisons. Cytokine Growth Factor Rev. 2003, 14, 297–309. [Google Scholar] [CrossRef]
- Regidor, D.L.; Detels, R.; Breen, E.C.; Widney, D.P.; Jacobson, L.P.; Palella, F.; Rinaldo, C.R.; Bream, J.H.; Martinez-Maza, O. Effect of highly active antiretroviral therapy on biomarkers of B-lymphocyte activation and inflammation. AIDS 2011, 25, 303–314. [Google Scholar] [CrossRef] [Green Version]
- Borges, A.H.; Silverberg, M.J.; Wentworth, D.; Grulich, A.E.; Fatkenheuer, G.; Mitsuyasu, R.; Tambussi, G.; Sabin, C.A.; Neaton, J.D.; Lundgren, J.D.; et al. Predicting risk of cancer during HIV infection: The role of inflammatory and coagulation biomarkers. AIDS 2013, 27, 1433–1441. [Google Scholar] [CrossRef] [Green Version]
- Vendrame, E.; Hussain, S.K.; Breen, E.C.; Magpantay, L.I.; Widney, D.P.; Jacobson, L.P.; Variakojis, D.; Knowlton, E.R.; Bream, J.H.; Ambinder, R.F.; et al. Serum levels of cytokines and biomarkers for inflammation and immune activation, and HIV-associated non-Hodgkin B-cell lymphoma risk. Cancer Epidemiol. Biomark. Prev. 2014, 23, 343–349. [Google Scholar] [CrossRef] [Green Version]
- Breen, E.C.; Hussain, S.K.; Magpantay, L.; Jacobson, L.P.; Detels, R.; Rabkin, C.S.; Kaslow, R.A.; Variakojis, D.; Bream, J.H.; Rinaldo, C.R.; et al. B-cell stimulatory cytokines and markers of immune activation are elevated several years prior to the diagnosis of systemic AIDS-associated non-Hodgkin B-cell lymphoma. Cancer Epidemiol. Biomark. Prev. 2011, 20, 1303–1314. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brenchley, J.M.; Schacker, T.W.; Ruff, L.E.; Price, D.A.; Taylor, J.H.; Beilman, G.J.; Nguyen, P.L.; Khoruts, A.; Larson, M.; Haase, A.T.; et al. CD4+ T cell depletion during all stages of HIV disease occurs predominantly in the gastrointestinal tract. J. Exp. Med. 2004, 200, 749–759. [Google Scholar] [CrossRef] [Green Version]
- Veazey, R.S.; DeMaria, M.; Chalifoux, L.V.; Shvetz, D.E.; Pauley, D.R.; Knight, H.L.; Rosenzweig, M.; Johnson, R.P.; Desrosiers, R.C.; Lackner, A.A. Gastrointestinal tract as a major site of CD4+ T cell depletion and viral replication in SIV infection. Science 1998, 280, 427–431. [Google Scholar] [CrossRef]
- Mehandru, S.; Poles, M.A.; Tenner-Racz, K.; Jean-Pierre, P.; Manuelli, V.; Lopez, P.; Shet, A.; Low, A.; Mohri, H.; Boden, D.; et al. Lack of mucosal immune reconstitution during prolonged treatment of acute and early HIV-1 infection. PLoS Med. 2006, 3, e484. [Google Scholar] [CrossRef]
- Tincati, C.; Biasin, M.; Bandera, A.; Violin, M.; Marchetti, G.; Piacentini, L.; Vago, G.L.; Balotta, C.; Moroni, M.; Franzetti, F.; et al. Early initiation of highly active antiretroviral therapy fails to reverse immunovirological abnormalities in gut-associated lymphoid tissue induced by acute HIV infection. Antivir. Ther. 2009, 14, 321–330. [Google Scholar]
- Nazli, A.; Chan, O.; Dobson-Belaire, W.N.; Ouellet, M.; Tremblay, M.J.; Gray-Owen, S.D.; Arsenault, A.L.; Kaushic, C. Exposure to HIV-1 directly impairs mucosal epithelial barrier integrity allowing microbial translocation. PLoS Pathog. 2010, 6, e1000852. [Google Scholar] [CrossRef]
- Klatt, N.R.; Estes, J.D.; Sun, X.; Ortiz, A.M.; Barber, J.S.; Harris, L.D.; Cervasi, B.; Yokomizo, L.K.; Pan, L.; Vinton, C.L.; et al. Loss of mucosal CD103+ DCs and IL-17+ and IL-22+ lymphocytes is associated with mucosal damage in SIV infection. Mucosal Immunol. 2012, 5, 646–657. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ryan, E.S.; Micci, L.; Fromentin, R.; Paganini, S.; McGary, C.S.; Easley, K.; Chomont, N.; Paiardini, M. Loss of Function of Intestinal IL-17 and IL-22 Producing Cells Contributes to Inflammation and Viral Persistence in SIV-Infected Rhesus Macaques. PLoS Pathog. 2016, 12, e1005412. [Google Scholar] [CrossRef]
- Kim, C.J.; Nazli, A.; Rojas, O.L.; Chege, D.; Alidina, Z.; Huibner, S.; Mujib, S.; Benko, E.; Kovacs, C.; Shin, L.Y.; et al. A role for mucosal IL-22 production and Th22 cells in HIV-associated mucosal immunopathogenesis. Mucosal Immunol. 2012, 5, 670–680. [Google Scholar] [CrossRef]
- Sandler, N.G.; Douek, D.C. Microbial translocation in HIV infection: Causes, consequences and treatment opportunities. Nat. Rev. Microbiol. 2012, 10, 655–666. [Google Scholar] [CrossRef]
- Mehraj, V.; Ramendra, R.; Isnard, S.; Dupuy, F.P.; Ponte, R.; Chen, J.; Kema, I.; Jenabian, M.A.; Costinuik, C.T.; Lebouche, B.; et al. Circulating (1-->3)-beta-D-glucan Is Associated with Immune Activation During Human Immunodeficiency Virus Infection. Clin. Infect. Dis. 2020, 70, 232–241. [Google Scholar] [CrossRef]
- Clifford, G.M.; Polesel, J.; Rickenbach, M.; Dal Maso, L.; Keiser, O.; Kofler, A.; Rapiti, E.; Levi, F.; Jundt, G.; Fisch, T.; et al. Cancer risk in the Swiss HIV Cohort Study: Associations with immunodeficiency, smoking, and highly active antiretroviral therapy. J. Natl. Cancer Inst. 2005, 97, 425–432. [Google Scholar] [CrossRef] [Green Version]
- Biggar, R.J.; Jaffe, E.S.; Goedert, J.J.; Chaturvedi, A.; Pfeiffer, R.; Engels, E.A. Hodgkin lymphoma and immunodeficiency in persons with HIV/AIDS. Blood 2006, 108, 3786–3791. [Google Scholar] [CrossRef] [PubMed]
- Lindqvist, M.; van Lunzen, J.; Soghoian, D.Z.; Kuhl, B.D.; Ranasinghe, S.; Kranias, G.; Flanders, M.D.; Cutler, S.; Yudanin, N.; Muller, M.I.; et al. Expansion of HIV-specific T follicular helper cells in chronic HIV infection. J. Clin. Investig. 2012, 122, 3271–3280. [Google Scholar] [CrossRef] [PubMed]
- Lorenzo-Redondo, R.; Fryer, H.R.; Bedford, T.; Kim, E.Y.; Archer, J.; Pond, S.L.K.; Chung, Y.S.; Penugonda, S.; Chipman, J.; Fletcher, C.V.; et al. Persistent HIV-1 replication maintains the tissue reservoir during therapy. Nature 2016, 530, 51–56. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Perreau, M.; Savoye, A.L.; De Crignis, E.; Corpataux, J.M.; Cubas, R.; Haddad, E.K.; De Leval, L.; Graziosi, C.; Pantaleo, G. Follicular helper T cells serve as the major CD4 T cell compartment for HIV-1 infection, replication, and production. J. Exp. Med. 2013, 210, 143–156. [Google Scholar] [CrossRef] [PubMed]
- Cubas, R.A.; Mudd, J.C.; Savoye, A.L.; Perreau, M.; van Grevenynghe, J.; Metcalf, T.; Connick, E.; Meditz, A.; Freeman, G.J.; Abesada-Terk, G., Jr.; et al. Inadequate T follicular cell help impairs B cell immunity during HIV infection. Nat. Med. 2013, 19, 494–499. [Google Scholar] [CrossRef] [Green Version]
- De Milito, A.; Morch, C.; Sonnerborg, A.; Chiodi, F. Loss of memory (CD27) B lymphocytes in HIV-1 infection. AIDS 2001, 15, 957–964. [Google Scholar] [CrossRef]
- Van Grevenynghe, J.; Cubas, R.A.; Noto, A.; DaFonseca, S.; He, Z.; Peretz, Y.; Filali-Mouhim, A.; Dupuy, F.P.; Procopio, F.A.; Chomont, N.; et al. Loss of memory B cells during chronic HIV infection is driven by Foxo3a- and TRAIL-mediated apoptosis. J. Clin. Investig. 2011, 121, 3877–3888. [Google Scholar] [CrossRef] [PubMed]
- Fenwick, C.; Joo, V.; Jacquier, P.; Noto, A.; Banga, R.; Perreau, M.; Pantaleo, G. T-cell exhaustion in HIV infection. Immunol. Rev. 2019, 292, 149–163. [Google Scholar] [CrossRef]
- Wherry, E.J.; Kurachi, M. Molecular and cellular insights into T cell exhaustion. Nat. Rev. Immunol. 2015, 15, 486–499. [Google Scholar] [CrossRef]
- Deeks, S.G. HIV infection, inflammation, immunosenescence, and aging. Annu. Rev. Med. 2011, 62, 141–155. [Google Scholar] [CrossRef] [Green Version]
- Allen, J.C.; Toapanta, F.R.; Chen, W.; Tennant, S.M. Understanding immunosenescence and its impact on vaccination of older adults. Vaccine 2020, 38, 8264–8272. [Google Scholar] [CrossRef]
- Clerici, M.; Stocks, N.I.; Zajac, R.A.; Boswell, R.N.; Lucey, D.R.; Via, C.S.; Shearer, G.M. Detection of three distinct patterns of T helper cell dysfunction in asymptomatic, human immunodeficiency virus-seropositive patients. Independence of CD4+ cell numbers and clinical staging. J. Clin. Investig. 1989, 84, 1892–1899. [Google Scholar] [CrossRef]
- Heather, J.M.; Best, K.; Oakes, T.; Gray, E.R.; Roe, J.K.; Thomas, N.; Friedman, N.; Noursadeghi, M.; Chain, B. Dynamic Perturbations of the T-Cell Receptor Repertoire in Chronic HIV Infection and following Antiretroviral Therapy. Front. Immunol. 2015, 6, 644. [Google Scholar] [CrossRef] [Green Version]
- Hernandez, D.M.; Valderrama, S.; Gualtero, S.; Hernandez, C.; Lopez, M.; Herrera, M.V.; Solano, J.; Fiorentino, S.; Quijano, S. Loss of T-Cell Multifunctionality and TCR-Vbeta Repertoire Against Epstein-Barr Virus Is Associated With Worse Prognosis and Clinical Parameters in HIV(+) Patients. Front. Immunol. 2018, 9, 2291. [Google Scholar] [CrossRef] [Green Version]
- Fromentin, R.; Bakeman, W.; Lawani, M.B.; Khoury, G.; Hartogensis, W.; DaFonseca, S.; Killian, M.; Epling, L.; Hoh, R.; Sinclair, E.; et al. CD4+ T Cells Expressing PD-1, TIGIT and LAG-3 Contribute to HIV Persistence during ART. PLoS Pathog. 2016, 12, e1005761. [Google Scholar] [CrossRef]
- Evans, V.A.; van der Sluis, R.M.; Solomon, A.; Dantanarayana, A.; McNeil, C.; Garsia, R.; Palmer, S.; Fromentin, R.; Chomont, N.; Sekaly, R.P.; et al. Programmed cell death-1 contributes to the establishment and maintenance of HIV-1 latency. AIDS 2018, 32, 1491–1497. [Google Scholar] [CrossRef]
- Henrich, T.J.; Hobbs, K.S.; Hanhauser, E.; Scully, E.; Hogan, L.E.; Robles, Y.P.; Leadabrand, K.S.; Marty, F.M.; Palmer, C.D.; Jost, S.; et al. Human Immunodeficiency Virus Type 1 Persistence Following Systemic Chemotherapy for Malignancy. J. Infect. Dis. 2017, 216, 254–262. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Slyker, J.A.; Guthrie, B.; Pankau, M.; Tapia, K.; Wamalwa, D.; Benki-Nugent, S.; Ngugi, E.; Huang, M.L.; Njuguna, I.; Langat, A.; et al. Cytomegalovirus and Epstein-Barr virus viremia are associated with HIV DNA levels in the reservoir of Kenyan infants on antiretroviral therapy. J. Infect. Dis. 2020. [Google Scholar] [CrossRef] [PubMed]
- Gianella, S.; Moser, C.; Vitomirov, A.; McKhann, A.; Layman, L.; Scott, B.; Caballero, G.; Lada, S.; Bosch, R.J.; Hoenigl, M.; et al. Presence of asymptomatic cytomegalovirus and Epstein-Barr virus DNA in blood of persons with HIV starting antiretroviral therapy is associated with non-AIDS clinical events. AIDS 2020, 34, 849–857. [Google Scholar] [CrossRef] [PubMed]
- Righetti, E.; Ballon, G.; Ometto, L.; Cattelan, A.M.; Menin, C.; Zanchetta, M.; Chieco-Bianchi, L.; De Rossi, A. Dynamics of Epstein-Barr virus in HIV-1-infected subjects on highly active antiretroviral therapy. AIDS 2002, 16, 63–73. [Google Scholar] [CrossRef]
- Basso, M.; Andreis, S.; Scaggiante, R.; Franchin, E.; Zago, D.; Biasolo, M.A.; Del Vecchio, C.; Mengoli, C.; Sarmati, L.; Andreoni, M.; et al. Cytomegalovirus, Epstein-Barr virus and human herpesvirus 8 salivary shedding in HIV positive men who have sex with men with controlled and uncontrolled plasma HIV viremia: A 24-month longitudinal study. BMC Infect. Dis. 2018, 18, 683. [Google Scholar] [CrossRef] [PubMed]
- Giron, L.B.; Ramos da Silva, S.; Barbosa, A.N.; Monteiro de Barros Almeida, R.A.; Rosario de Souza, L.; Elgui de Oliveira, D. Impact of Epstein-Barr virus load, virus genotype, and frequency of the 30 bp deletion in the viral BNLF-1 gene in patients harboring the human immunodeficiency virus. J. Med. Virol. 2013, 85, 2110–2118. [Google Scholar] [CrossRef] [PubMed]
- De Franca, T.R.; de Albuquerque Tavares Carvalho, A.; Gomes, V.B.; Gueiros, L.A.; Porter, S.R.; Leao, J.C. Salivary shedding of Epstein-Barr virus and cytomegalovirus in people infected or not by human immunodeficiency virus 1. Clin. Oral Investig. 2012, 16, 659–664. [Google Scholar] [CrossRef]
- Lisco, A.; Munawwar, A.; Introini, A.; Vanpouille, C.; Saba, E.; Feng, X.; Grivel, J.C.; Singh, S.; Margolis, L. Semen of HIV-1-infected individuals: Local shedding of herpesviruses and reprogrammed cytokine network. J. Infect. Dis. 2012, 205, 97–105. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gianella, S.; Morris, S.R.; Vargas, M.V.; Young, J.A.; Callahan, B.; Richman, D.D.; Little, S.J.; Smith, D.M. Role of seminal shedding of herpesviruses in HIV Type 1 Transmission. J. Infect. Dis. 2013, 207, 257–261. [Google Scholar] [CrossRef]
- Gantt, S.; Carlsson, J.; Shetty, A.K.; Seidel, K.D.; Qin, X.; Mutsvangwa, J.; Musingwini, G.; Woelk, G.; Zijenah, L.S.; Katzenstein, D.A.; et al. Cytomegalovirus and Epstein-Barr virus in breast milk are associated with HIV-1 shedding but not with mastitis. AIDS 2008, 22, 1453–1460. [Google Scholar] [CrossRef] [Green Version]
- Moriuchi, M.; Moriuchi, H. Increased susceptibility to HIV-1 of peripheral blood lymphocytes in acute infection with Epstein-Barr virus. J. Med. Virol. 2003, 71, 343–346. [Google Scholar] [CrossRef] [PubMed]
- Guan, M.; Zhang, R.D.; Wu, B.; Henderson, E.E. Infection of primary CD4+ and CD8+ T lymphocytes by Epstein-Barr virus enhances human immunodeficiency virus expression. J. Virol. 1996, 70, 7341–7346. [Google Scholar] [CrossRef] [Green Version]
- Lauener, R.P.; Huttner, S.; Buisson, M.; Hossle, J.P.; Albisetti, M.; Seigneurin, J.M.; Seger, R.A.; Nadal, D. T-cell death by apoptosis in vertically human immunodeficiency virus-infected children coincides with expansion of CD8+/interleukin-2 receptor-/HLA-DR+ T cells: Sign of a possible role for herpes viruses as cofactors? Blood 1995, 86, 1400–1407. [Google Scholar] [CrossRef]
- Quintana, M.D.P.; Smith-Togobo, C.; Moormann, A.; Hviid, L. Endemic Burkitt lymphoma—An aggressive childhood cancer linked to Plasmodium falciparum exposure, but not to exposure to other malaria parasites. APMIS 2020, 128, 129–135. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sebina, I.; Fogg, L.G.; James, K.R.; Soon, M.S.F.; Akter, J.; Thomas, B.S.; Hill, G.R.; Engwerda, C.R.; Haque, A. IL-6 promotes CD4(+) T-cell and B-cell activation during Plasmodium infection. Parasite Immunol. 2017, 39. [Google Scholar] [CrossRef]
- Alves, F.A.; Pelajo-Machado, M.; Totino, P.R.; Souza, M.T.; Goncalves, E.C.; Schneider, M.P.; Muniz, J.A.; Krieger, M.A.; Andrade, M.C.; Daniel-Ribeiro, C.T.; et al. Splenic architecture disruption and parasite-induced splenocyte activation and anergy in Plasmodium falciparum-infected Saimiri sciureus monkeys. Malar. J. 2015, 14, 128. [Google Scholar] [CrossRef] [Green Version]
- Wilmore, J.R.; Asito, A.S.; Wei, C.; Piriou, E.; Sumba, P.O.; Sanz, I.; Rochford, R. AID expression in peripheral blood of children living in a malaria holoendemic region is associated with changes in B cell subsets and Epstein-Barr virus. Int. J. Cancer 2015, 136, 1371–1380. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Torgbor, C.; Awuah, P.; Deitsch, K.; Kalantari, P.; Duca, K.A.; Thorley-Lawson, D.A. A multifactorial role for P. falciparum malaria in endemic Burkitt’s lymphoma pathogenesis. PLoS Pathog. 2014, 10, e1004170. [Google Scholar] [CrossRef] [Green Version]
- Robbiani, D.F.; Deroubaix, S.; Feldhahn, N.; Oliveira, T.Y.; Callen, E.; Wang, Q.; Jankovic, M.; Silva, I.T.; Rommel, P.C.; Bosque, D.; et al. Plasmodium Infection Promotes Genomic Instability and AID-Dependent B Cell Lymphoma. Cell 2015, 162, 727–737. [Google Scholar] [CrossRef] [Green Version]
- Ramiro, A.R.; Jankovic, M.; Eisenreich, T.; Difilippantonio, S.; Chen-Kiang, S.; Muramatsu, M.; Honjo, T.; Nussenzweig, A.; Nussenzweig, M.C. AID is required for c-myc/IgH chromosome translocations in vivo. Cell 2004, 118, 431–438. [Google Scholar] [CrossRef] [Green Version]
- Donati, D.; Zhang, L.P.; Chene, A.; Chen, Q.; Flick, K.; Nystrom, M.; Wahlgren, M.; Bejarano, M.T. Identification of a polyclonal B-cell activator in Plasmodium falciparum. Infect. Immun. 2004, 72, 5412–5418. [Google Scholar] [CrossRef] [Green Version]
- Donati, D.; Mok, B.; Chene, A.; Xu, H.; Thangarajh, M.; Glas, R.; Chen, Q.; Wahlgren, M.; Bejarano, M.T. Increased B cell survival and preferential activation of the memory compartment by a malaria polyclonal B cell activator. J. Immunol. 2006, 177, 3035–3044. [Google Scholar] [CrossRef]
- Chene, A.; Donati, D.; Guerreiro-Cacais, A.O.; Levitsky, V.; Chen, Q.; Falk, K.I.; Orem, J.; Kironde, F.; Wahlgren, M.; Bejarano, M.T. A molecular link between malaria and Epstein-Barr virus reactivation. PLoS Pathog. 2007, 3, e80. [Google Scholar] [CrossRef] [PubMed]
- Frech, C.; Chen, N. Genome comparison of human and non-human malaria parasites reveals species subset-specific genes potentially linked to human disease. PLoS Comput. Biol. 2011, 7, e1002320. [Google Scholar] [CrossRef]
- Mulama, D.H.; Bailey, J.A.; Foley, J.; Chelimo, K.; Ouma, C.; Jura, W.G.; Otieno, J.; Vulule, J.; Moormann, A.M. Sickle cell trait is not associated with endemic Burkitt lymphoma: An ethnicity and malaria endemicity-matched case-control study suggests factors controlling EBV may serve as a predictive biomarker for this pediatric cancer. Int. J. Cancer 2014, 134, 645–653. [Google Scholar] [CrossRef] [Green Version]
- Hviid, L. Naturally acquired immunity to Plasmodium falciparum malaria in Africa. Acta Trop. 2005, 95, 270–275. [Google Scholar] [CrossRef] [PubMed]
- Burkitt, D.; Wright, D. Geographical and tribal distribution of the African lymphoma in Uganda. Br. Med. J. 1966, 1, 569–573. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, I.; Clarke, S.E.; Gosling, R.; Hamainza, B.; Killeen, G.; Magill, A.; O’Meara, W.; Price, R.N.; Riley, E.M. “Asymptomatic” Malaria: A Chronic and Debilitating Infection That Should Be Treated. PLoS Med. 2016, 13, e1001942. [Google Scholar] [CrossRef]
- Karimi, P.; Birmann, B.M.; Anderson, L.A.; McShane, C.M.; Gadalla, S.M.; Sampson, J.N.; Mbulaiteye, S.M. Risk factors for Burkitt lymphoma: A nested case-control study in the UK Clinical Practice Research Datalink. Br. J. Haematol. 2018, 181, 505–514. [Google Scholar] [CrossRef] [PubMed]
- Moormann, A.M.; Chelimo, K.; Sumba, O.P.; Lutzke, M.L.; Ploutz-Snyder, R.; Newton, D.; Kazura, J.; Rochford, R. Exposure to holoendemic malaria results in elevated Epstein-Barr virus loads in children. J. Infect. Dis. 2005, 191, 1233–1238. [Google Scholar] [CrossRef] [Green Version]
- Njie, R.; Bell, A.I.; Jia, H.; Croom-Carter, D.; Chaganti, S.; Hislop, A.D.; Whittle, H.; Rickinson, A.B. The effects of acute malaria on Epstein-Barr virus (EBV) load and EBV-specific T cell immunity in Gambian children. J. Infect. Dis. 2009, 199, 31–38. [Google Scholar] [CrossRef] [Green Version]
- Reynaldi, A.; Schlub, T.E.; Chelimo, K.; Sumba, P.O.; Piriou, E.; Ogolla, S.; Moormann, A.M.; Rochford, R.; Davenport, M.P. Impact of Plasmodium falciparum Coinfection on Longitudinal Epstein-Barr Virus Kinetics in Kenyan Children. J. Infect. Dis. 2016, 213, 985–991. [Google Scholar] [CrossRef] [Green Version]
- Chattopadhyay, P.K.; Chelimo, K.; Embury, P.B.; Mulama, D.H.; Sumba, P.O.; Gostick, E.; Ladell, K.; Brodie, T.M.; Vulule, J.; Roederer, M.; et al. Holoendemic malaria exposure is associated with altered Epstein-Barr virus-specific CD8(+) T-cell differentiation. J. Virol. 2013, 87, 1779–1788. [Google Scholar] [CrossRef] [Green Version]
- Snider, C.J.; Cole, S.R.; Chelimo, K.; Sumba, P.O.; Macdonald, P.D.; John, C.C.; Meshnick, S.R.; Moormann, A.M. Recurrent Plasmodium falciparum malaria infections in Kenyan children diminish T-cell immunity to Epstein Barr virus lytic but not latent antigens. PLoS ONE 2012, 7, e31753. [Google Scholar] [CrossRef]
- Moormann, A.M.; Heller, K.N.; Chelimo, K.; Embury, P.; Ploutz-Snyder, R.; Otieno, J.A.; Oduor, M.; Munz, C.; Rochford, R. Children with endemic Burkitt lymphoma are deficient in EBNA1-specific IFN-gamma T cell responses. Int. J. Cancer 2009, 124, 1721–1726. [Google Scholar] [CrossRef] [Green Version]
- Parsons, E.; Otieno, J.A.; Ong’echa, J.M.; Nixon, C.E.; Vulule, J.; Munz, C.; Stewart, V.A.; Moormann, A.M. Regulatory T Cells in Endemic Burkitt Lymphoma Patients Are Associated with Poor Outcomes: A Prospective, Longitudinal Study. PLoS ONE 2016, 11, e0167841. [Google Scholar] [CrossRef] [Green Version]
- Forconi, C.S.; Cosgrove, C.P.; Saikumar-Lakshmi, P.; Nixon, C.E.; Foley, J.; Ong’echa, J.M.; Otieno, J.A.; Alter, G.; Munz, C.; Moormann, A.M. Poorly cytotoxic terminally differentiated CD56(neg)CD16(pos) NK cells accumulate in Kenyan children with Burkitt lymphomas. Blood Adv. 2018, 2, 1101–1114. [Google Scholar] [CrossRef] [Green Version]
- Kato, A.; Imai, K.; Ochiai, K.; Ogata, Y. Prevalence and quantitative analysis of Epstein-Barr virus DNA and Porphyromonas gingivalis associated with Japanese chronic periodontitis patients. Clin. Oral Investig. 2015, 19, 1605–1610. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sunde, P.T.; Olsen, I.; Enersen, M.; Grinde, B. Patient with severe periodontitis and subgingival Epstein-Barr virus treated with antiviral therapy. J. Clin. Virol. 2008, 42, 176–178. [Google Scholar] [CrossRef]
- Slots, J. Periodontal herpesviruses: Prevalence, pathogenicity, systemic risk. Periodontol. 2000 2015, 69, 28–45. [Google Scholar] [CrossRef]
- Slots, J.; Slots, H. Periodontal herpesvirus morbidity and treatment. Periodontol. 2000 2019, 79, 210–220. [Google Scholar] [CrossRef]
- Miller, C.S.; Avdiushko, S.A.; Kryscio, R.J.; Danaher, R.J.; Jacob, R.J. Effect of prophylactic valacyclovir on the presence of human herpesvirus DNA in saliva of healthy individuals after dental treatment. J. Clin. Microbiol. 2005, 43, 2173–2180. [Google Scholar] [CrossRef] [Green Version]
- Kikuchi, K.; Noguchi, Y.; de Rivera, M.W.; Hoshino, M.; Sakashita, H.; Yamada, T.; Inoue, H.; Miyazaki, Y.; Nozaki, T.; Gonzalez-Lopez, B.S.; et al. Detection of Epstein-Barr virus genome and latent infection gene expression in normal epithelia, epithelial dysplasia, and squamous cell carcinoma of the oral cavity. Tumour Biol. 2016, 37, 3389–3404. [Google Scholar] [CrossRef]
- Vincent-Bugnas, S.; Vitale, S.; Mouline, C.C.; Khaali, W.; Charbit, Y.; Mahler, P.; Precheur, I.; Hofman, P.; Maryanski, J.L.; Doglio, A. EBV infection is common in gingival epithelial cells of the periodontium and worsens during chronic periodontitis. PLoS ONE 2013, 8, e80336. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Imai, K.; Ogata, Y. How Does Epstein-Barr Virus Contribute to Chronic Periodontitis? Int. J. Mol. Sci. 2020, 21, 1940. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Michalowicz, B.S.; Ronderos, M.; Camara-Silva, R.; Contreras, A.; Slots, J. Human herpesviruses and Porphyromonas gingivalis are associated with juvenile periodontitis. J. Periodontol. 2000, 71, 981–988. [Google Scholar] [CrossRef]
- Elamin, A.; Ali, R.W.; Bakken, V. Putative periodontopathic bacteria and herpes viruses interactions in the subgingival plaque of patients with aggressive periodontitis and healthy controls. Clin. Exp. Dent. Res. 2017, 3, 183–190. [Google Scholar] [CrossRef]
- Chen, C.; Feng, P.; Slots, J. Herpesvirus-bacteria synergistic interaction in periodontitis. Periodontol. 2000 2020, 82, 42–64. [Google Scholar] [CrossRef]
- Hadinoto, V.; Shapiro, M.; Sun, C.C.; Thorley-Lawson, D.A. The dynamics of EBV shedding implicate a central role for epithelial cells in amplifying viral output. PLoS Pathog. 2009, 5, e1000496. [Google Scholar] [CrossRef] [Green Version]
- Countryman, J.K.; Gradoville, L.; Miller, G. Histone hyperacetylation occurs on promoters of lytic cycle regulatory genes in Epstein-Barr virus-infected cell lines which are refractory to disruption of latency by histone deacetylase inhibitors. J. Virol. 2008, 82, 4706–4719. [Google Scholar] [CrossRef] [Green Version]
- Ghosh, S.K.; Perrine, S.P.; Williams, R.M.; Faller, D.V. Histone deacetylase inhibitors are potent inducers of gene expression in latent EBV and sensitize lymphoma cells to nucleoside antiviral agents. Blood 2012, 119, 1008–1017. [Google Scholar] [CrossRef] [Green Version]
- Makino, K.; Takeichi, O.; Imai, K.; Inoue, H.; Hatori, K.; Himi, K.; Saito, I.; Ochiai, K.; Ogiso, B. Porphyromonas endodontalis reactivates latent Epstein-Barr virus. Int. Endod. J. 2018, 51, 1410–1419. [Google Scholar] [CrossRef] [PubMed]
- Imai, K.; Inoue, H.; Tamura, M.; Cueno, M.E.; Inoue, H.; Takeichi, O.; Kusama, K.; Saito, I.; Ochiai, K. The periodontal pathogen Porphyromonas gingivalis induces the Epstein-Barr virus lytic switch transactivator ZEBRA by histone modification. Biochimie 2012, 94, 839–846. [Google Scholar] [CrossRef] [PubMed]
- Niederman, R.; Buyle-Bodin, Y.; Lu, B.Y.; Robinson, P.; Naleway, C. Short-chain carboxylic acid concentration in human gingival crevicular fluid. J. Dent. Res. 1997, 76, 575–579. [Google Scholar] [CrossRef] [PubMed]
- Koike, R.; Nodomi, K.; Watanabe, N.; Ogata, Y.; Takeichi, O.; Takei, M.; Kaneko, T.; Tonogi, M.; Kotani, A.I.; Imai, K. Butyric Acid in Saliva of Chronic Periodontitis Patients Induces Transcription of the EBV Lytic Switch Activator BZLF1: A Pilot Study. In Vivo 2020, 34, 587–594. [Google Scholar] [CrossRef] [Green Version]
- Imai, K.; Yamada, K.; Tamura, M.; Ochiai, K.; Okamoto, T. Reactivation of latent HIV-1 by a wide variety of butyric acid-producing bacteria. Cell. Mol. Life Sci. 2012, 69, 2583–2592. [Google Scholar] [CrossRef] [PubMed]
- Desai, S.N.; Landay, A.L. HIV and aging: Role of the microbiome. Curr. Opin. HIV AIDS 2018, 13, 22–27. [Google Scholar] [CrossRef] [PubMed]
- Stern, J.; Shai, E.; Zaks, B.; Halabi, A.; Houri-Haddad, Y.; Shapira, L.; Palmon, A. Reduced expression of gamma interferon in serum and marked lymphoid depletion induced by Porphyromonas gingivalis increase murine morbidity and mortality due to cytomegalovirus infection. Infect. Immun. 2004, 72, 5791–5798. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rush, M.C.; Simon, M.W. Occurrence of Epstein-Barr virus illness in children diagnosed with group A streptococcal pharyngitis. Clin. Pediatr. 2003, 42, 417–420. [Google Scholar] [CrossRef]
- Schlecht, N.F.; Platt, R.W.; Duarte-Franco, E.; Costa, M.C.; Sobrinho, J.P.; Prado, J.C.; Ferenczy, A.; Rohan, T.E.; Villa, L.L.; Franco, E.L. Human papillomavirus infection and time to progression and regression of cervical intraepithelial neoplasia. J. Natl. Cancer Inst. 2003, 95, 1336–1343. [Google Scholar] [CrossRef] [Green Version]
- De Lima, M.A.P.; Neto, P.J.N.; Lima, L.P.M.; Goncalves Junior, J.; Teixeira Junior, A.G.; Teodoro, I.P.P.; Facundo, H.T.; da Silva, C.G.L.; Lima, M.V.A. Association between Epstein-Barr virus (EBV) and cervical carcinoma: A meta-analysis. Gynecol. Oncol. 2018, 148, 317–328. [Google Scholar] [CrossRef]
- Khenchouche, A.; Sadouki, N.; Boudriche, A.; Houali, K.; Graba, A.; Ooka, T.; Bouguermouh, A. Human papillomavirus and Epstein-Barr virus co-infection in cervical carcinoma in Algerian women. Virol. J. 2013, 10, 340. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Se Thoe, S.Y.; Wong, K.K.; Pathmanathan, R.; Sam, C.K.; Cheng, H.M.; Prasad, U. Elevated secretory IgA antibodies to Epstein-Barr virus (EBV) and presence of EBV DNA and EBV receptors in patients with cervical carcinoma. Gynecol. Oncol. 1993, 50, 168–172. [Google Scholar] [CrossRef]
- Landers, R.J.; O’Leary, J.J.; Crowley, M.; Healy, I.; Annis, P.; Burke, L.; O’Brien, D.; Hogan, J.; Kealy, W.F.; Lewis, F.A.; et al. Epstein-Barr virus in normal, pre-malignant, and malignant lesions of the uterine cervix. J. Clin. Pathol. 1993, 46, 931–935. [Google Scholar] [CrossRef] [Green Version]
- Sasagawa, T.; Shimakage, M.; Nakamura, M.; Sakaike, J.; Ishikawa, H.; Inoue, M. Epstein-Barr virus (EBV) genes expression in cervical intraepithelial neoplasia and invasive cervical cancer: A comparative study with human papillomavirus (HPV) infection. Hum. Pathol. 2000, 31, 318–326. [Google Scholar] [CrossRef]
- Shimakage, M.; Sasagawa, T. Detection of Epstein-Barr virus-determined nuclear antigen-2 mRNA by in situ hybridization. J. Virol. Methods 2001, 93, 23–32. [Google Scholar] [CrossRef]
- Al-Thawadi, H.; Ghabreau, L.; Aboulkassim, T.; Yasmeen, A.; Vranic, S.; Batist, G.; Al Moustafa, A.E. Co-Incidence of Epstein-Barr Virus and High-Risk Human Papillomaviruses in Cervical Cancer of Syrian Women. Front. Oncol. 2018, 8, 250. [Google Scholar] [CrossRef] [Green Version]
- Aromseree, S.; Pientong, C.; Swangphon, P.; Chaiwongkot, A.; Patarapadungkit, N.; Kleebkaow, P.; Tungsiriwattana, T.; Kongyingyoes, B.; Vendrig, T.; Middeldorp, J.M.; et al. Possible contributing role of Epstein-Barr virus (EBV) as a cofactor in human papillomavirus (HPV)-associated cervical carcinogenesis. J. Clin. Virol. 2015, 73, 70–76. [Google Scholar] [CrossRef]
- Hilton, D.A.; Brown, L.J.; Pringle, J.H.; Nandha, H. Absence of Epstein-Barr virus in carcinoma of the cervix. Cancer 1993, 72, 1946–1948. [Google Scholar] [CrossRef]
- Shoji, Y.; Saegusa, M.; Takano, Y.; Hashimura, M.; Okayasu, I. Detection of the Epstein-Barr virus genome in cervical neoplasia is closely related to the degree of infiltrating lymphoid cells: A polymerase chain reaction and in situ hybridization approach. Pathol. Int. 1997, 47, 507–511. [Google Scholar] [CrossRef]
- Elgui de Oliveira, D.; Furtado Monteiro, T.A.; Alencar de Melo, W.; Amaral Reboucas Moreira, M.; Alvarenga, M.; Bacchi, C.E. Lack of Epstein-Barr virus infection in cervical carcinomas. Arch. Pathol. Lab. Med. 1999, 123, 1098–1100. [Google Scholar] [CrossRef]
- Yang, Y.Y.; Koh, L.W.; Tsai, J.H.; Tsai, C.H.; Wong, E.F.; Lin, S.J.; Yang, C.C. Correlation of viral factors with cervical cancer in Taiwan. J. Microbiol. Immunol. Infect. 2004, 37, 282–287. [Google Scholar]
- Seo, S.S.; Kim, W.H.; Song, Y.S.; Kim, S.H.; Kim, J.W.; Park, N.H.; Kang, S.B.; Lee, H.P. Epstein-Barr virus plays little role in cervical carcinogenesis in Korean women. Int. J. Gynecol. Cancer. 2005, 15, 312–318. [Google Scholar] [CrossRef]
- Blanco, R.; Carrillo-Beltran, D.; Osorio, J.C.; Calaf, G.M.; Aguayo, F. Role of Epstein-Barr Virus and Human Papillomavirus Coinfection in Cervical Cancer: Epidemiology, Mechanisms and Perspectives. Pathogens 2020, 9, 685. [Google Scholar] [CrossRef]
- Kitano, Y.; Fujisaki, S.; Nakamura, N.; Miyazaki, K.; Katsuki, T.; Okamura, H. Immunological disorder against the Epstein-Barr virus infection and prognosis in patients with cervical carcinoma. Gynecol. Oncol. 1995, 57, 150–157. [Google Scholar] [CrossRef]
- Kahla, S.; Oueslati, S.; Achour, M.; Kochbati, L.; Chanoufi, M.B.; Maalej, M.; Oueslati, R. Correlation between ebv co-infection and HPV16 genome integrity in Tunisian cervical cancer patients. Braz. J. Microbiol. 2012, 43, 744–753. [Google Scholar] [CrossRef] [Green Version]
- Szostek, S.; Zawilinska, B.; Kopec, J.; Kosz-Vnenchak, M. Herpesviruses as possible cofactors in HPV-16-related oncogenesis. Acta Biochim. Pol. 2009, 56, 337–342. [Google Scholar] [CrossRef] [Green Version]
- McCormick, T.M.; Canedo, N.H.; Furtado, Y.L.; Silveira, F.A.; de Lima, R.J.; Rosman, A.D.; Almeida Filho, G.L.; Carvalho Mda, G. Association between human papillomavirus and Epstein—Barr virus DNA and gene promoter methylation of RB1 and CDH1 in the cervical lesions: A transversal study. Diagn. Pathol. 2015, 10, 59. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Termine, N.; Panzarella, V.; Falaschini, S.; Russo, A.; Matranga, D.; Lo Muzio, L.; Campisi, G. HPV in oral squamous cell carcinoma vs head and neck squamous cell carcinoma biopsies: A meta-analysis (1988–2007). Ann. Oncol. 2008, 19, 1681–1690. [Google Scholar] [CrossRef]
- Staff, P.O. Correction: Epstein-Barr virus and human papillomavirus infections and genotype distribution in head and neck cancers. PLoS ONE 2015, 10, e0118439. [Google Scholar] [CrossRef] [Green Version]
- Laantri, N.; Attaleb, M.; Kandil, M.; Naji, F.; Mouttaki, T.; Dardari, R.; Belghmi, K.; Benchakroun, N.; El Mzibri, M.; Khyatti, M. Human papillomavirus detection in moroccan patients with nasopharyngeal carcinoma. Infect. Agent Cancer 2011, 6, 3. [Google Scholar] [CrossRef] [Green Version]
- Mirzamani, N.; Salehian, P.; Farhadi, M.; Tehran, E.A. Detection of EBV and HPV in nasopharyngeal carcinoma by in situ hybridization. Exp. Mol. Pathol. 2006, 81, 231–234. [Google Scholar] [CrossRef] [PubMed]
- Tung, Y.C.; Lin, K.H.; Chu, P.Y.; Hsu, C.C.; Kuo, W.R. Detection of human papilloma virus and Epstein-Barr virus DNA in nasopharyngeal carcinoma by polymerase chain reaction. Kaohsiung J. Med. Sci. 1999, 15, 256–262. [Google Scholar] [PubMed]
- Lin, Z.; Khong, B.; Kwok, S.; Cao, H.; West, R.B.; Le, Q.T.; Kong, C.S. Human papillomavirus 16 detected in nasopharyngeal carcinomas in white Americans but not in endemic Southern Chinese patients. Head Neck 2014, 36, 709–714. [Google Scholar] [CrossRef]
- Stenmark, M.H.; McHugh, J.B.; Schipper, M.; Walline, H.M.; Komarck, C.; Feng, F.Y.; Worden, F.P.; Wolf, G.T.; Chepeha, D.B.; Prince, M.E.; et al. Nonendemic HPV-positive nasopharyngeal carcinoma: Association with poor prognosis. Int. J. Radiat. Oncol. Biol. Phys. 2014, 88, 580–588. [Google Scholar] [CrossRef] [Green Version]
- Dogan, S.; Hedberg, M.L.; Ferris, R.L.; Rath, T.J.; Assaad, A.M.; Chiosea, S.I. Human papillomavirus and Epstein-Barr virus in nasopharyngeal carcinoma in a low-incidence population. Head Neck 2014, 36, 511–516. [Google Scholar] [CrossRef] [PubMed]
- Maxwell, J.H.; Kumar, B.; Feng, F.Y.; McHugh, J.B.; Cordell, K.G.; Eisbruch, A.; Worden, F.P.; Wolf, G.T.; Prince, M.E.; Moyer, J.S.; et al. HPV-positive/p16-positive/EBV-negative nasopharyngeal carcinoma in white North Americans. Head Neck 2010, 32, 562–567. [Google Scholar] [CrossRef]
- Robinson, M.; Suh, Y.E.; Paleri, V.; Devlin, D.; Ayaz, B.; Pertl, L.; Thavaraj, S. Oncogenic human papillomavirus-associated nasopharyngeal carcinoma: An observational study of correlation with ethnicity, histological subtype and outcome in a UK population. Infect Agent Cancer 2013, 8, 30. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, R.; Ekshyyan, O.; Moore-Medlin, T.; Rong, X.; Nathan, S.; Gu, X.; Abreo, F.; Rosenthal, E.L.; Shi, M.; Guidry, J.T.; et al. Association between human papilloma virus/Epstein-Barr virus coinfection and oral carcinogenesis. J. Oral Pathol. Med. 2015, 44, 28–36. [Google Scholar] [CrossRef] [PubMed]
- Nawandar, D.M.; Ohashi, M.; Djavadian, R.; Barlow, E.; Makielski, K.; Ali, A.; Lee, D.; Lambert, P.F.; Johannsen, E.; Kenney, S.C. Differentiation-Dependent LMP1 Expression Is Required for Efficient Lytic Epstein-Barr Virus Reactivation in Epithelial Cells. J. Virol. 2017, 91. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- White, E.A. Manipulation of Epithelial Differentiation by HPV Oncoproteins. Viruses 2019, 11, 369. [Google Scholar] [CrossRef] [Green Version]
- Scholle, F.; Bendt, K.M.; Raab-Traub, N. Epstein-Barr virus LMP2A transforms epithelial cells, inhibits cell differentiation, and activates Akt. J. Virol. 2000, 74, 10681–10689. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guidry, J.T.; Myers, J.E.; Bienkowska-Haba, M.; Songock, W.K.; Ma, X.; Shi, M.; Nathan, C.O.; Bodily, J.M.; Sapp, M.J.; Scott, R.S. Inhibition of Epstein-Barr Virus Replication in Human Papillomavirus-Immortalized Keratinocytes. J. Virol. 2019, 93. [Google Scholar] [CrossRef] [Green Version]
- Makielski, K.R.; Lee, D.; Lorenz, L.D.; Nawandar, D.M.; Chiu, Y.F.; Kenney, S.C.; Lambert, P.F. Human papillomavirus promotes Epstein-Barr virus maintenance and lytic reactivation in immortalized oral keratinocytes. Virology 2016, 495, 52–62. [Google Scholar] [CrossRef] [PubMed]
- Nawandar, D.M.; Wang, A.; Makielski, K.; Lee, D.; Ma, S.; Barlow, E.; Reusch, J.; Jiang, R.; Wille, C.K.; Greenspan, D.; et al. Differentiation-Dependent KLF4 Expression Promotes Lytic Epstein-Barr Virus Infection in Epithelial Cells. PLoS Pathog. 2015, 11, e1005195. [Google Scholar] [CrossRef]
- De Lima, M.A.P.; Teodoro, I.P.P.; Galiza, L.E.; Filho, P.; Marques, F.M.; Pinheiro Junior, R.F.F.; Macedo, G.E.C.; Facundo, H.T.; da Silva, C.G.L.; Lima, M.V.A. Association between Epstein-Barr Virus and Oral Carcinoma: A Systematic Review with Meta-Analysis. Crit. Rev. Oncog. 2019, 24, 349–368. [Google Scholar] [CrossRef] [PubMed]
- Shimabuku, T.; Tamanaha, A.; Kitamura, B.; Tanabe, Y.; Tawata, N.; Ikehara, F.; Arakaki, K.; Kinjo, T. Dual expression of Epstein-Barr virus, latent membrane protein-1 and human papillomavirus-16 E6 transform primary mouse embryonic fibroblasts through NF-kappaB signaling. Int. J. Clin. Exp. Pathol. 2014, 7, 1920–1934. [Google Scholar] [PubMed]
- Murphy, G.; Pfeiffer, R.; Camargo, M.C.; Rabkin, C.S. Meta-analysis shows that prevalence of Epstein-Barr virus-positive gastric cancer differs based on sex and anatomic location. Gastroenterology 2009, 137, 824–833. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Davila-Collado, R.; Jarquin-Duran, O.; Dong, L.T.; Espinoza, J.L. Epstein-Barr Virus and Helicobacter pylori Co-Infection in Non-Malignant Gastroduodenal Disorders. Pathogens 2020, 9, 104. [Google Scholar] [CrossRef] [Green Version]
- Saxena, A.; Nath Prasad, K.; Chand Ghoshal, U.; Krishnani, N.; Roshan Bhagat, M.; Husain, N. Association of Helicobacter pylori and Epstein-Barr virus with gastric cancer and peptic ulcer disease. Scand. J. Gastroenterol. 2008, 43, 669–674. [Google Scholar] [CrossRef]
- Castaneda, C.A.; Castillo, M.; Chavez, I.; Barreda, F.; Suarez, N.; Nieves, J.; Bernabe, L.A.; Valdivia, D.; Ruiz, E.; Dias-Neto, E.; et al. Prevalence of Helicobacter pylori Infection, Its Virulent Genotypes, and Epstein-Barr Virus in Peruvian Patients with Chronic Gastritis and Gastric Cancer. J. Glob. Oncol. 2019, 5, 1–9. [Google Scholar] [CrossRef]
- Shukla, S.K.; Prasad, K.N.; Tripathi, A.; Ghoshal, U.C.; Krishnani, N.; Husain, N. Expression profile of latent and lytic transcripts of epstein-barr virus in patients with gastroduodenal diseases: A study from northern India. J. Med. Virol. 2012, 84, 1289–1297. [Google Scholar] [CrossRef]
- Del Moral-Hernandez, O.; Castanon-Sanchez, C.A.; Reyes-Navarrete, S.; Martinez-Carrillo, D.N.; Betancourt-Linares, R.; Jimenez-Wences, H.; de la Pena, S.; Roman-Roman, A.; Hernandez-Sotelo, D.; Fernandez-Tilapa, G. Multiple infections by EBV, HCMV and Helicobacter pylori are highly frequent in patients with chronic gastritis and gastric cancer from Southwest Mexico: An observational study. Medicine 2019, 98, e14124. [Google Scholar] [CrossRef]
- Shukla, S.K.; Prasad, K.N.; Tripathi, A.; Singh, A.; Saxena, A.; Ghoshal, U.C.; Krishnani, N.; Husain, N. Epstein-Barr virus DNA load and its association with Helicobacter pylori infection in gastroduodenal diseases. Braz. J. Infect. Dis. 2011, 15, 583–590. [Google Scholar] [CrossRef] [Green Version]
- De Souza, C.R.; de Oliveira, K.S.; Ferraz, J.J.; Leal, M.F.; Calcagno, D.Q.; Seabra, A.D.; Khayat, A.S.; Montenegro, R.C.; Alves, A.P.; Assumpcao, P.P.; et al. Occurrence of Helicobacter pylori and Epstein-Barr virus infection in endoscopic and gastric cancer patients from Northern Brazil. BMC Gastroenterol. 2014, 14, 179. [Google Scholar] [CrossRef] [Green Version]
- De Souza, C.R.T.; Almeida, M.C.A.; Khayat, A.S.; da Silva, E.L.; Soares, P.C.; Chaves, L.C.; Burbano, R.M.R. Association between Helicobacter pylori, Epstein-Barr virus, human papillomavirus and gastric adenocarcinomas. World J. Gastroenterol. 2018, 24, 4928–4938. [Google Scholar] [CrossRef]
- Cardenas-Mondragon, M.G.; Torres, J.; Sanchez-Zauco, N.; Gomez-Delgado, A.; Camorlinga-Ponce, M.; Maldonado-Bernal, C.; Fuentes-Panana, E.M. Elevated Levels of Interferon-gamma Are Associated with High Levels of Epstein-Barr Virus Reactivation in Patients with the Intestinal Type of Gastric Cancer. J. Immunol. Res. 2017, 2017, 7069242. [Google Scholar] [CrossRef] [Green Version]
- Cardenas-Mondragon, M.G.; Torres, J.; Flores-Luna, L.; Camorlinga-Ponce, M.; Carreon-Talavera, R.; Gomez-Delgado, A.; Kasamatsu, E.; Fuentes-Panana, E.M. Case-control study of Epstein-Barr virus and Helicobacter pylori serology in Latin American patients with gastric disease. Br. J. Cancer 2015, 112, 1866–1873. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cardenas-Mondragon, M.G.; Carreon-Talavera, R.; Camorlinga-Ponce, M.; Gomez-Delgado, A.; Torres, J.; Fuentes-Panana, E.M. Epstein Barr virus and Helicobacter pylori co-infection are positively associated with severe gastritis in pediatric patients. PLoS ONE 2013, 8, e62850. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Minoura-Etoh, J.; Gotoh, K.; Sato, R.; Ogata, M.; Kaku, N.; Fujioka, T.; Nishizono, A. Helicobacter pylori-associated oxidant monochloramine induces reactivation of Epstein-Barr virus (EBV) in gastric epithelial cells latently infected with EBV. J. Med. Microbiol. 2006, 55, 905–911. [Google Scholar] [CrossRef] [Green Version]
- Pandey, S.; Jha, H.C.; Shukla, S.K.; Shirley, M.K.; Robertson, E.S. Epigenetic Regulation of Tumor Suppressors by Helicobacter pylori Enhances EBV-Induced Proliferation of Gastric Epithelial Cells. mBio 2018, 9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ansari, S.; Yamaoka, Y. Helicobacter pylori Virulence Factor Cytotoxin-Associated Gene A (CagA)-Mediated Gastric Pathogenicity. Int. J. Mol. Sci. 2020, 21, 7430. [Google Scholar] [CrossRef] [PubMed]
- Saju, P.; Murata-Kamiya, N.; Hayashi, T.; Senda, Y.; Nagase, L.; Noda, S.; Matsusaka, K.; Funata, S.; Kunita, A.; Urabe, M.; et al. Host SHP1 phosphatase antagonizes Helicobacter pylori CagA and can be downregulated by Epstein-Barr virus. Nat. Microbiol. 2016, 1, 16026. [Google Scholar] [CrossRef]
- Olofsson, A.; Vallstrom, A.; Petzold, K.; Tegtmeyer, N.; Schleucher, J.; Carlsson, S.; Haas, R.; Backert, S.; Wai, S.N.; Grobner, G.; et al. Biochemical and functional characterization of Helicobacter pylori vesicles. Mol. Microbiol. 2010, 77, 1539–1555. [Google Scholar] [CrossRef] [PubMed] [Green Version]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sánchez-Ponce, Y.; Fuentes-Pananá, E.M. The Role of Coinfections in the EBV–Host Broken Equilibrium. Viruses 2021, 13, 1399. https://0-doi-org.brum.beds.ac.uk/10.3390/v13071399
Sánchez-Ponce Y, Fuentes-Pananá EM. The Role of Coinfections in the EBV–Host Broken Equilibrium. Viruses. 2021; 13(7):1399. https://0-doi-org.brum.beds.ac.uk/10.3390/v13071399
Chicago/Turabian StyleSánchez-Ponce, Yessica, and Ezequiel M. Fuentes-Pananá. 2021. "The Role of Coinfections in the EBV–Host Broken Equilibrium" Viruses 13, no. 7: 1399. https://0-doi-org.brum.beds.ac.uk/10.3390/v13071399