Dengue — Quo tu et quo vadis?
Abstract
:1. Introduction
2. History, Origin and Emergence of DENV
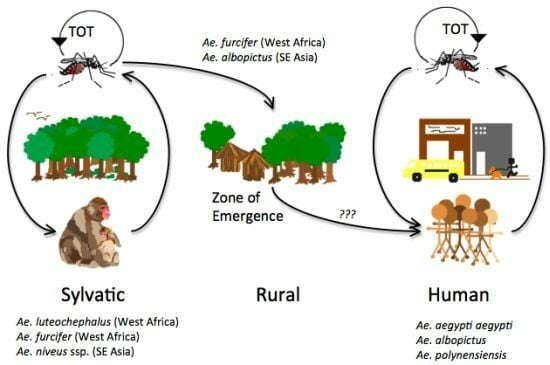
3. Transmission Cycles
4. Phylogeny as a Tool to Understand DENV Epidemiology and Evolution
4.1. DENV-1

4.2. DENV-2

4.3. DENV-3


4.4. DENV-4

5. Basis of Genetic Diversity of DENV
6. Evolution Pattern and its Driving Forces
6.1. Dengue Evolution is Characterized by Frequent Lineage Replacement
6.2. The Role of Selection Pressures on Shaping the Evolution and Population Dynamics of DENV
6.3. Cross-Immunity as a Driving Force for DENV Evolution
6.4. Stochastic Events Play an Important Role in Clade Replacement
6.5. Antibody-Dependent Enhancement (ADE) and Its Effect on DENV Evolution
7. DENV Evolutionary Rates and Their Constraints
8. Conclusions
Acknowledgments
Conflict of Interest
References and Notes
- Calisher, C.H.; Karabatsos, N.; Dalrymple, J.M.; Shope, R.E.; Porterfield, J.S.; Westaway, E.G.; Brandt, W.E. Antigenic relationships between flaviviruses as determined by cross-neutralization tests with polyclonal antisera. J. Gen. Virol. 1989, 70, 37–43. [Google Scholar] [CrossRef]
- Rudnick, A. Dengue virus ecology in Malaysia. Inst. Med. Res. Malays. Bull. 1986, 23, 51–152. [Google Scholar]
- Cornet, M.; Saluzzo, J.F.; Hervy, J.P.; Digoutte, J.P.; Germain, M.; Chauvancy, M.F.; Eyraud, M.; Ferrara, L.; Heme, G.; Legros, F. Dengue 2 au Senegal oriental: Une poussee epizootique en milieu selvatique; isolements du virus a partir des moustiques et d’un singe et considerations epidemiologiques. Cah. ORSTOM. ser Ent. Med. et Parasitol. 1984, 22, 313–323. [Google Scholar]
- de Thoisy, B.; Dussart, P.; Kazanji, M. Wild terrestrial rainforest mammals as potential reservoirs for flaviviruses (yellow fever, dengue 2 and St Louis encephalitis viruses) in French Guiana. Trans. R. Soc. Trop. Med. Hyg. 2004, 98, 409–412. [Google Scholar] [CrossRef]
- de Thoisy, B.; Lacoste, V.; Germain, A.; Munoz-Jordan, J.; Colon, C.; Mauffrey, J.F.; Delaval, M.; Catzeflis, F.; Kazanji, M.; Matheus, S.; et al. Dengue infection in neotropical forest mammals. Vector Borne Zoonotic Dis. 2009, 9, 157–170. [Google Scholar] [CrossRef] [PubMed]
- Rodhain, F. The role of monkeys in the biology of dengue and yellow fever. Comp. Immunol. Microbiol. Infect. Dis. 1991, 14, 9–19. [Google Scholar] [CrossRef] [PubMed]
- Gubler, D.J. Dengue and dengue hemorrhagic fever: Its history and resurgence as a global public health problem. In Dengue and Dengue Hemorrhagic Fever; Gubler, D.J., Kuno, G., Eds.; CABI Publishing: Oxon, UK, 1997; pp. 1–22. [Google Scholar]
- Guzman, M.G.; Halstead, S.B.; Artsob, H.; Buchy, P.; Farrar, J.; Gubler, D.J.; Hunsperger, E.; Kroeger, A.; Margolis, H.S.; Martinez, E.; et al. Dengue: A continuing global threat. Nat. Rev. Microbiol. 2010, 8, S7–S16. [Google Scholar] [CrossRef] [PubMed]
- Natiello, M.; Ritacco, V.; Morales, M.A.; Deodato, B.; Picollo, M.; Dinerstein, E.; Enria, D. Indigenous dengue fever, Buenos Aires, Argentina. Emerg. Infect. Dis. 2008, 14, 1498–1499. [Google Scholar] [CrossRef] [PubMed]
- CDC. Locally acquired dengue—Key West, Florida, 2009–2010. MMWR 2010, 59, 577–581. [Google Scholar]
- Kyle, J.L.; Harris, E. Global spread and persistence of dengue. Ann. Rev. Microbiol. 2008, 62, 71–92. [Google Scholar] [CrossRef]
- Kalayanarooj, S.; Nimmannitya, S. Clinical presentations of dengue hemorrhagic fever in infants compared to children. J. Med. Assoc. Thailand 2003, 86, S673–S680. [Google Scholar]
- Kongsomboon, K.; Singhasivanon, P.; Kaewkungwal, J.; Nimmannitya, S.; Mammen, M.P., Jr.; Nisalak, A.; Sawanpanyalert, P. Temporal trends of dengue fever/dengue hemorrhagic fever in Bangkok, Thailand from 1981 to 2000: An age-period-cohort analysis. Southeast Asian J. Trop. Med. Publ. Health 2004, 35, 913–917. [Google Scholar]
- Witayathawornwong, P. DHF in infants, late infants and older children: A comparative study. Southeast Asian J. Trop. Med. Publ. Health 2005, 36, 896–900. [Google Scholar]
- Guilarde, A.O.; Turchi, M.D.; Siqueira, J.B., Jr.; Feres, V.C.; Rocha, B.; Levi, J.E.; Souza, V.A.; Boas, L.S.; Pannuti, C.S.; Martelli, C.M. Dengue and dengue hemorrhagic fever among adults: Clinical outcomes related to viremia, serotypes, and antibody response. J. Infect. Dis. 2008, 197, 817–824. [Google Scholar] [CrossRef]
- Hanafusa, S.; Chanyasanha, C.; Sujirarat, D.; Khuankhunsathid, I.; Yaguchi, A.; Suzuki, T. Clinical features and differences between child and adult dengue infections in Rayong province, southeast Thailand. Southeast Asian J. Trop. Med. Publ. Health 2008, 39, 252–259. [Google Scholar]
- Koh, B.K.; Ng, L.C.; Kita, Y.; Tang, C.S.; Ang, L.W.; Wong, K.Y.; James, L.; Goh, K.T. The 2005 dengue epidemic in Singapore: Epidemiology, prevention and control. Ann. Acad. Med. Singapore 2008, 37, 538–545. [Google Scholar] [CrossRef]
- Lee, I.K.; Liu, J.W.; Yang, K.D. Clinical and laboratory characteristics and risk factors for fatality in elderly patients with dengue hemorrhagic fever. Am. J. Trop. Med. Hyg. 2008, 79, 149–153. [Google Scholar] [CrossRef] [PubMed]
- Siqueira, J.B., Jr.; Martelli, C.M.; Coelho, G.E.; Simplicio, A.C.; Hatch, D.L. Dengue and dengue hemorrhagic fever, Brazil, 1981–2002. Emerg. Infect. Dis. 2005, 11, 48–53. [Google Scholar] [CrossRef]
- Wichmann, O.; Hongsiriwon, S.; Bowonwatanuwong, C.; Chotivanich, K.; Sukthana, Y.; Pukrittayakamee, S. Risk factors and clinical features associated with severe dengue infection in adults and children during the 2001 epidemic in Chonburi, Thailand. Trop. Med. Int. Health 2004, 9, 1022–1029. [Google Scholar] [CrossRef]
- Leo, Y.S.; Thein, T.L.; Fisher, D.A.; Low, J.G.; Oh, H.M.; Narayanan, R.L.; Gan, V.C.; Lee, V.J.; Lye, D.C. Confirmed adult dengue deaths in Singapore: 5-year multi-center retrospective study. BMC Infect. Dis. 2011, 11, 123. [Google Scholar] [CrossRef]
- Fox, A.; Le, N.M.; Simmons, C.P.; Wolbers, M.; Wertheim, H.F.; Pham, T.K.; Tran, T.H.; Trinh, T.M.; Nguyen, T.L.; Nguyen, V.T.; et al. Immunological and viral determinants of dengue severity in hospitalized adults in Ha Noi, Viet Nam. PLoS Negl. Trop. Dis. 2011, 5, e967. [Google Scholar] [CrossRef] [PubMed]
- Tee, H.P.; How, S.H.; Jamalludin, A.R.; Safhan, M.N.; Sapian, M.M.; Kuan, Y.C.; Sapari, S. Risk factors associated with development of dengue haemorrhagic fever or dengue shock syndrome in adults in hospital Tengku Ampuan Afzan Kuantan. Med. J. Malaysia 2009, 64, 316–320. [Google Scholar] [PubMed]
- Halstead, S.B.; Nimmannitya, S.; Cohen, S.N. Observations related to pathogenesis of dengue hemorrhagic fever. IV. Relation of disease severity to antibody response and virus recovered. Yale J. Biol. Med. 1970, 42, 311–328. [Google Scholar]
- Kliks, S.C.; Nimmanitya, S.; Nisalak, A.; Burke, D.S. Evidence that maternal dengue antibodies are important in the development of dengue hemorrhagic fever in infants. Am. J. Trop. Med. Hyg. 1988, 38, 411–419. [Google Scholar] [CrossRef] [PubMed]
- Dejnirattisai, W.; Jumnainsong, A.; Onsirisakul, N.; Fitton, P.; Vasanawathana, S.; Limpitikul, W.; Puttikhunt, C.; Edwards, C.; Duangchinda, T.; Supasa, S.; Chawansuntati, K.; Malasit, P.; Mongkolsapaya, J.; Screaton, G. Cross-reacting antibodies enhance dengue virus infection in humans. Science 2010, 328, 745–748. [Google Scholar] [CrossRef]
- Vaughn, D.W.; Green, S.; Kalayanarooj, S.; Innis, B.L.; Nimmannitya, S.; Suntayakorn, S.; Endy, T.P.; Raengsakulrach, B.; Rothman, A.L.; Ennis, F.A.; Nisalak, A. Dengue viremia titer, antibody response pattern, and virus serotype correlate with disease severity. J. Infect. Dis. 2000, 181, 2–9. [Google Scholar] [CrossRef]
- Wang, W.K.; Chao, D.Y.; Kao, C.L.; Wu, H.C.; Liu, Y.C.; Li, C.M.; Lin, S.C.; Ho, S.T.; Huang, J.H.; King, C.C. High levels of plasma dengue viral load during defervescence in patients with dengue hemorrhagic fever: Implications for pathogenesis. Virology 2003, 305, 330–338. [Google Scholar] [CrossRef]
- Araujo, J.M.; de Filippis, A.M.; Schatzmayr, H.G.; de Araujo, E.S.; Britto, C.; Cardoso, M.A.; Camacho, L.A.; Nogueira, R.M. Quantification of dengue virus type 3 RNA in fatal and non-fatal cases in Brazil, 2002. Trans. R. Soc. Trop. Med. Hyg. 2009, 103, 952–954. [Google Scholar] [CrossRef]
- Anders, K.L.; Nguyet, N.M.; Chau, N.V.; Hung, N.T.; Thuy, T.T.; Lien le, B.; Farrar, J.; Wills, B.; Hien, T.T.; Simmons, C.P. Epidemiological factors associated with dengue shock syndrome and mortality in hospitalized dengue patients in Ho Chi Minh City, Vietnam. Am. J. Trop. Med. Hyg. 2011, 84, 127–134. [Google Scholar] [CrossRef]
- Guzman, M.G.; Kouri, G.; Morier, L.; Soler, M.; Fernandez, A. A study of fatal hemorrhagic dengue cases in Cuba, 1981. Bull. Pan Am. Health Organ. 1984, 18, 213–220. [Google Scholar]
- Kalayanarooj, S.; Nimmannitya, S. Is dengue severity related to nutritional status? Southeast Asian J. Trop. Med. Publ. Health 2005, 36, 378–384. [Google Scholar]
- Thisyakorn, U.; Nimmannitya, S. Nutritional status of children with dengue hemorrhagic fever. Clin. Infect. Dis. 1993, 16, 295–297. [Google Scholar] [CrossRef] [PubMed]
- Bravo, J.R.; Guzman, M.G.; Kouri, G.P. Why dengue haemorrhagic fever in Cuba? 1. Individual risk factors for dengue haemorrhagic fever/dengue shock syndrome (DHF/DSS). Trans. R. Soc. Trop. Med. Hyg. 1987, 81, 816–820. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.P.; Kikuchi, M.; Vu, T.Q.; Do, Q.H.; Tran, T.T.; Vo, D.T.; Ha, M.T.; Vo, V.T.; Cao, T.P.; Tran, V.D.; et al. Protective and enhancing HLA alleles, HLA-DRB1*0901 and HLA-A*24, for severe forms of dengue virus infection, dengue hemorrhagic fever and dengue shock syndrome. PLoS Negl. Trop. Dis. 2008, 2, e304. [Google Scholar]
- Appanna, R.; Ponnampalavanar, S.; Lum Chai See, L.; Sekaran, S.D. Susceptible and protective HLA class 1 alleles against dengue fever and dengue hemorrhagic fever patients in a Malaysian population. PLoS ONE 2010, 5, e13029. [Google Scholar] [CrossRef] [PubMed]
- Beatty, M.E.; Beutels, P.; Meltzer, M.I.; Shepard, D.S.; Hombach, J.; Hutubessy, R.; Dessis, D.; Coudeville, L.; Dervaux, B.; Wichmann, O.; et al. Health economics of dengue: A systematic literature review and expert panel’s assessment. Am. J. Trop. Med. Hyg. 2011, 84, 473–488. [Google Scholar] [CrossRef]
- Beaute, J.; Vong, S. Cost and disease burden of dengue in Cambodia. BMC Publ. Health 2010, 10, 521. [Google Scholar] [CrossRef]
- Luz, P.M.; Grinsztejn, B.; Galvani, A.P. Disability adjusted life years lost to dengue in Brazil. Trop. Med. Int. Health 2009, 14, 237–246. [Google Scholar] [CrossRef]
- Anderson, K.B.; Chunsuttiwat, S.; Nisalak, A.; Mammen, M.P.; Libraty, D.H.; Rothman, A.L.; Green, S.; Vaughn, D.W.; Ennis, F.A.; Endy, T.P. Burden of symptomatic dengue infection in children at primary school in Thailand: A prospective study. Lancet 2007, 369, 1452–1459. [Google Scholar] [CrossRef]
- Cho Min, N. Assessment of dengue hemorrhagic fever in Myanmar. Southeast Asian J. Trop. Med. Publ. Health 2000, 31, 636–641. [Google Scholar]
- Meltzer, M.I.; Rigau-Perez, J.G.; Clark, G.G.; Reiter, P.; Gubler, D.J. Using disability-adjusted life years to assess the economic impact of dengue in Puerto Rico: 1984–1994. Am. J. Trop. Med. Hyg. 1998, 59, 265–271. [Google Scholar] [CrossRef] [PubMed]
- Gubler, D.J.; Meltzer, M. Impact of dengue/dengue hemorrhagic fever on the developing world. Adv. Virus Res. 1999, 53, 35–70. [Google Scholar] [PubMed]
- Clark, D.V.; Mammen, M.P., Jr.; Nisalak, A.; Puthimethee, V.; Endy, T.P. Economic impact of dengue fever/dengue hemorrhagic fever in Thailand at the family and population levels. Am. J. Trop. Med. Hyg. 2005, 72, 786–791. [Google Scholar] [CrossRef]
- Garg, P.; Nagpal, J.; Khairnar, P.; Seneviratne, S.L. Economic burden of dengue infections in India. Trans. R. Soc. Trop. Med. Hyg. 2008, 102, 570–577. [Google Scholar] [CrossRef] [PubMed]
- Shepard, D.S.; Coudeville, L.; Halasa, Y.A.; Zambrano, B.; Dayan, G.H. Economic impact of dengue illness in the Americas. Am. J. Trop. Med. Hyg. 2011, 84, 200–207. [Google Scholar] [CrossRef] [PubMed]
- Huy, R.; Wichmann, O.; Beatty, M.; Ngan, C.; Duong, S.; Margolis, H.S.; Vong, S. Cost of dengue and other febrile illnesses to households in rural Cambodia: A prospective community-based case-control study. BMC Publ. Health 2009, 9, 155. [Google Scholar] [CrossRef]
- Suaya, J.A.; Shepard, D.S.; Siqueira, J.B.; Martelli, C.T.; Lum, L.C.; Tan, L.H.; Kongsin, S.; Jiamton, S.; Garrido, F.; Montoya, R.; et al. Cost of dengue cases in eight countries in the Americas and Asia: A prospective study. Am. J. Trop. Med. Hyg. 2009, 80, 846–855. [Google Scholar] [CrossRef]
- Armien, B.; Suaya, J.A.; Quiroz, E.; Sah, B.K.; Bayard, V.; Marchena, L.; Campos, C.; Shepard, D.S. Clinical characteristics and national economic cost of the 2005 dengue epidemic in Panama. Am. J. Trop. Med. Hyg. 2008, 79, 364–371. [Google Scholar] [CrossRef]
- Harving, M.L.; Ronsholt, F.F. The economic impact of dengue hemorrhagic fever on family level in southern Vietnam. Danish Med. Bull. 2007, 54, 170–172. [Google Scholar]
- Anez, G.; Balza, R.; Valero, N.; Larreal, Y. Economic impact of dengue and dengue hemorrhagic fever in the state of Zulia, Venezuela, 1997–2003. Rev. Panamericana Salud Publ. 2006, 19, 314–320. [Google Scholar]
- Valdes, L.G.; Mizhrahi, J.V.; Guzman, M.G. Economic impact of dengue 2 epidemic in Santiago de Cuba, 1997. Rev. Cubana Med. Trop. 2002, 54, 220–227. [Google Scholar] [PubMed]
- Leung, A. Diseases of the premodern period in china. In The Cambridge World History of Human Disease; Kilple, K.F., Ed.; Cambridge University Press: Cambridge, UK, 1993; pp. 354–361. [Google Scholar]
- Bylon, D. Korte aantekening, wegens eene algemeene ziekte, doorgaans genaamd de knokkel-koorts. In Verhandelungen van het bataviaasch genootschop der konsten in wetenschappen; Johannes Allart (Amsterdam): Batavia, 1780; pp. 17–30. [Google Scholar]
- Christie, J. On epidemics of dengue fever: Their diffusion and etiology. Glasgow Med. J. 1881, 16, 161–176. [Google Scholar] [PubMed]
- Hirsch, A. Dengue, a comparatively new desease: Its symptoms. In Handbook of geographical and historical pathology; Syndenham Society: London, 1883; Volume I, pp. 55–81. [Google Scholar]
- Rush, A.B. An account of the bilious remitting fever, as it appeared in Philadelphia in the summer and autumn of the year 1780. In Medical Inquiries and Observations; Prichard and Hall: Philadelphia, PA, USA, 1789; pp. 89–100. [Google Scholar]
- Vasilakis, N.; Weaver, S.C. The history and evolution of human dengue emergence. Adv. Vir. Res. 2008, 72, 1–76. [Google Scholar]
- Pepper, P. A note on David Bylon and dengue. Ann. Med. Hist. 1941, 3, 363–368. [Google Scholar] [PubMed]
- Carey, D.E. Chikungunya and dengue: A case of mistaken identity? J. Hist. Med. Allied Sci. 1971, 26, 243–262. [Google Scholar] [CrossRef]
- Simmons, J.S.; St John, J.H.; Reynolds, F.H.K. Experimental studies of dengue. Philippine J. Sci. 1931, 44, 1–252. [Google Scholar]
- Rico-Hesse, R. Molecular evolution and distribution of dengue viruses type 1 and 2 in nature. Virology 1990, 174, 479–493. [Google Scholar] [CrossRef]
- Wang, E.; Ni, H.; Xu, R.; Barrett, A.D.; Watowich, S.J.; Gubler, D.J.; Weaver, S.C. Evolutionary relationships of endemic/epidemic and sylvatic dengue viruses. J. Virol. 2000, 74, 3227–3234. [Google Scholar] [CrossRef]
- Vasilakis, N.; Hanley, K.A.; Weaver, S.C. Dengue virus emergence from its sylvatic cycle. In Frontiers in Dengue Virus Research; Hanley, K.A., Weaver, S.C., Eds.; Caister Academic Press: Norfork, UK, 2010; pp. 183–220. [Google Scholar]
- Twiddy, S.S.; Farrar, J.J.; Vinh Chau, N.; Wills, B.; Gould, E.A.; Gritsun, T.; Lloyd, G.; Holmes, E.C. Phylogenetic relationships and differential selection pressures among genotypes of dengue-2 virus. Virology 2002, 298, 63–72. [Google Scholar] [CrossRef]
- Twiddy, S.S.; Woelk, C.H.; Holmes, E.C. Phylogenetic evidence for adaptive evolution of dengue viruses in nature. J. Gen. Virol. 2002, 83, 1679–1689. [Google Scholar] [CrossRef]
- Twiddy, S.S.; Holmes, E.C.; Rambaut, A. Inferring the rate and time-scale of dengue virus evolution. Mol. Biol. Evol. 2003, 20, 122–129. [Google Scholar] [CrossRef] [PubMed]
- Holmes, E.C.; Twiddy, S.S. The origin, emergence and evolutionary genetics of dengue virus. Infect. Genet. Evol. 2003, 3, 19–28. [Google Scholar] [CrossRef] [PubMed]
- Araujo, J.M.; Nogueira, R.M.; Schatzmayr, H.G.; Zanotto, P.M.; Bello, G. Phylogeography and evolutionary history of dengue virus type 3. Infect. Genet. Evol. 2009, 9, 716–725. [Google Scholar] [CrossRef]
- Villabona-Arenas, C.J.; Zanotto, P.M. Evolutionary history of dengue virus type 4: Insights into genotype phylodynamics. Infect. Genet. Evol. 2011, 11, 878–885. [Google Scholar] [CrossRef] [PubMed]
- Dunham, E.J.; Holmes, E.C. Inferring the timescale of dengue virus evolution under realistic models of DNA substitution. J. Mol. Evol. 2007, 64, 656–661. [Google Scholar] [CrossRef] [PubMed]
- Zanotto, P.M.; Gould, E.A.; Gao, G.F.; Harvey, P.H.; Holmes, E.C. Population dynamics of flaviviruses revealed by molecular phylogenies. Proc. Natl. Acad. Sci. U. S. A. 1996, 93, 548–553. [Google Scholar] [CrossRef] [PubMed]
- Vasilakis, N.; Durbin, A.; Travassos da Rosa, A.P.A.; Munoz-Jordan, J.L.; Tesh, R.B.; Weaver, S.C. Antigenic relationships between sylvatic and endemic dengue viruses. Am. J. Trop. Med. Hyg. 2008, 79, 128–132. [Google Scholar] [CrossRef]
- Sabin, A.B. Research on dengue during World War II. Am. J. Trop. Med. Hyg. 1952, 1, 30–50. [Google Scholar] [CrossRef]
- Ferguson, N.M.; Donnelly, C.A.; Anderson, R.M. Transmission dynamics and epidemiology of dengue: Insights from age-stratified sero-prevalence surveys. Phil. Trans. R. Soc. London 1999, 354, 757–768. [Google Scholar] [CrossRef]
- Ferguson, N.; Anderson, R.; Gupta, S. The effect of antibody-dependent enhancement on the transmission dynamics and persistence of multiple-strain pathogens. Proc. Natl. Acad. Sci. U. S. A. 1999, 96, 790–794. [Google Scholar] [CrossRef]
- Cologna, R.; Armstrong, P.M.; Rico-Hesse, R. Selection for virulent dengue viruses occurs in humans and mosquitoes. J. Virol. 2005, 79, 853–859. [Google Scholar] [CrossRef] [PubMed]
- Cologna, R.; Rico-Hesse, R. American genotype structures decrease dengue virus output from human monocytes and dendritic cells. J. Virol. 2003, 77, 3929–3938. [Google Scholar] [CrossRef] [PubMed]
- Cummings, D.A.; Irizarry, R.A.; Huang, N.E.; Endy, T.P.; Nisalak, A.; Ungchusak, K.; Burke, D.S. Travelling waves in the occurrence of dengue haemorrhagic fever in Thailand. Nature 2004, 427, 344–347. [Google Scholar] [CrossRef] [PubMed]
- Billings, L.; Schwartz, I.B.; Shaw, L.B.; McCrary, M.; Burke, D.S.; Cummings, D.A. Instabilities in multiserotype disease models with antibody-dependent enhancement. J. Theor. Biol. 2007, 246, 18–27. [Google Scholar] [CrossRef] [PubMed]
- Armstrong, P.M.; Rico-Hesse, R. Efficiency of dengue serotype 2 virus strains to infect and disseminate in Aedes aegypti. Am. J. Trop. Med. Hyg. 2003, 68, 539–544. [Google Scholar] [CrossRef]
- Armstrong, P.M.; Rico-Hesse, R. Differential susceptibility of Aedes aegypti to infection by the American and southeast Asian genotypes of dengue type 2 virus. Vector Borne Zoonotic Dis. 2001, 1, 159–168. [Google Scholar] [CrossRef] [PubMed]
- Messer, W.B.; Gubler, D.J.; Harris, E.; Sivananthan, K.; de Silva, A.M. Emergence and global spread of a dengue serotype 3, subtype III virus. Emerg. Infect. Dis. 2003, 9, 800–809. [Google Scholar] [CrossRef]
- Gaunt, M.W.; Sall, A.A.; de Lamballerie, X.; Falconar, A.K.; Dzhivanian, T.I.; Gould, E.A. Phylogenetic relationships of flaviviruses correlate with their epidemiology, disease association and biogeography. J. Gen. Virol. 2001, 82, 1867–1876. [Google Scholar] [CrossRef]
- Smith, C.E. The history of dengue in tropical Asia and its probable relationship to the mosquito Aedes aegypti. J. Trop. Med. Hyg. 1956, 59, 243–251. [Google Scholar]
- Bryant, J.E.; Holmes, E.C.; Barrett, A.D. Out of Africa: A molecular perspective on the introduction of yellow fever virus into the Americas. PLoS Pathog. 2007, 3, e75. [Google Scholar] [CrossRef]
- Vasconcelos, P.F.; Costa, Z.G.; Travassos Da Rosa, E.S.; Luna, E.; Rodrigues, S.G.; Barros, V.L.; Dias, J.P.; Monteiro, H.A.; Oliva, O.F.; Vasconcelos, H.B.; et al. Epidemic of jungle yellow fever in Brazil, 2000: Implications of climatic alterations in disease spread. J. Med. Virol. 2001, 65, 598–604. [Google Scholar] [CrossRef] [PubMed]
- Vasconcelos, P.F.; Rodrigues, S.G.; Degallier, N.; Moraes, M.A.; da Rosa, J.F.; da Rosa, E.S.; Mondet, B.; Barros, V.L.; da Rosa, A.P. An epidemic of sylvatic yellow fever in the southeast region of Maranhao state, Brazil, 1993–1994: Epidemiologic and entomologic findings. Am. J. Trop. Med. Hyg. 1997, 57, 132–137. [Google Scholar] [CrossRef] [PubMed]
- Tabachnick, W.J.; Powell, J.R. A world-wide survey of genetic variation in the yellow fever mosquito, Aedes aegypti. Genet. Res. 1979, 34, 215–229. [Google Scholar] [CrossRef] [PubMed]
- Powell, J.R.; Tabachnick, W.J.; Arnold, J. Genetics and the origin of a vector population: Aedes aegypti, a case study. Science 1980, 208, 1385–1387. [Google Scholar] [CrossRef] [PubMed]
- Diallo, M.; Sall, A.A.; Moncayo, A.C.; Ba, Y.; Fernandez, Z.; Ortiz, D.; Coffey, L.L.; Mathiot, C.; Tesh, R.B.; Weaver, S.C. Potential role of sylvatic and domestic african mosquito species in dengue emergence. Am. J. Trop. Med. Hyg. 2005, 73, 445–449. [Google Scholar] [CrossRef]
- Diallo, M.; Ba, Y.; Faye, O.; Soumare, M.L.; Dia, I.; Sall, A.A. Vector competence of aedes aegypti populations from Senegal for sylvatic and epidemic dengue 2 virus isolated in West Africa. Trans. R. Soc. Trop. Med. Hyg. 2008, 102, 493–498. [Google Scholar] [CrossRef]
- Black, W.C.; Bennett, K.E.; Gorrochotegui-Escalante, N.; Barillas-Mury, C.V.; Fernandez-Salas, I.; de Lourdes Munoz, M.; Farfan-Ale, J.A.; Olson, K.E.; Beaty, B.J. Flavivirus susceptibility in Aedes aegypti. Arch. Med. Res. 2002, 33, 379–388. [Google Scholar] [CrossRef]
- Vasilakis, N.; Cardosa, J.; Hanley, K.A.; Holmes, E.C.; Weaver, S.C. Fever from the forest: Prospects for the continued emergence of sylvatic dengue virus and its impact on public health. Nat. Rev. Microbiol. 2011, 9, 532–541. [Google Scholar] [CrossRef]
- Cordellier, R.; Bouchite, B.; Roche, J.C.; Monteny, N.; Diaco, B.; Akoliba, P. Circulation selvatique du virus dengue 2, en 1980, dans les savanes sub-soudaniennes de Cote d’Ivoire. Cah. ORSTOM. ser Ent. Med. et Parasitol. 1983, 21, 165–179. [Google Scholar]
- Roche, J.C.; Cordellier, R.; Hervy, J.P.; Digoutte, J.P.; Monteny, N. Isolement de 96 souches de virus dengue 2 a partir de moustiques captures en Cote d’Ivoire et en Haute Volta. Ann. Virol. (Institut Pasteur) 1983, 134E, 233–244. [Google Scholar] [CrossRef]
- Vasilakis, N.; Tesh, R.B.; Weaver, S.C. Sylvatic dengue virus type 2 activity in humans, Nigeria, 1966. Emerg. Infect. Dis. 2008, 14, 502–504. [Google Scholar] [CrossRef]
- Weaver, S.C.; Vasilakis, N. Molecular evolution of dengue viruses: Contributions of phylogenetics to understanding the history and epidemiology of the preeminent arboviral disease. Infect. Genet. Evol. 2009, 9, 523–540. [Google Scholar] [CrossRef] [PubMed]
- Lambrechts, L.; Scott, T.W.; Gubler, D.J. Consequences of the expanding global distribution of Aedes albopictus for dengue virus transmission. PLoS Negl. Trop. Dis. 2010, 4, e646. [Google Scholar] [CrossRef] [PubMed]
- Rosen, L.; Roseboom, L.E.; Gubler, D.J.; Lien, J.C.; Chaniotis, B.N. Comparative susceptibility of mosquito species and strains to oral and parenteral infection with dengue and Japanese encephalitis viruses. Am. J. Trop. Med. Hyg. 1985, 34, 603–615. [Google Scholar] [CrossRef] [PubMed]
- Leichtenstern, O. Influenza and dengue. In Specielle pathologie und therapie; Nothnagel, H., Ed.; Alfred Holder: Wien, Austria, 1896; Volume IV, pp. 133–226. [Google Scholar]
- Cleland, J.E.; Bradley, B.; McDonald, W. On the transmission of Australian dengue by the mosquito Stegomyia fasciata. Med. J. Aust. 1916, 2, 179–200. [Google Scholar] [CrossRef]
- Siler, J.F.; Hall, M.W.; Hitchens, A.P. Dengue: Its history, epidemiology, mechanism of transmission, etiology, clinical manifestations, immunity, and prevention. Philippine J. Sci. 1926, 29, 1–252. [Google Scholar]
- Pybus, O.G.; Rambaut, A.; Holmes, E.C.; Harvey, P.H. New inferences from tree shape: Numbers of missing taxa and population growth rates. Syst. Biol. 2002, 51, 881–888. [Google Scholar] [CrossRef]
- Diallo, M.; Ba, Y.; Sall, A.A.; Diop, O.M.; Ndione, J.A.; Mondo, M.; Girault, L.; Mathiot, C. Amplification of the sylvatic cycle of dengue virus type 2, Senegal, 1999–2000: Entomologic findings and epidemiologic considerations. Emerg. Infect. Dis. 2003, 9, 362–367. [Google Scholar] [CrossRef]
- Hervy, J.P.; Legros, F.; Roche, J.C.; Monteny, N.; Diaco, B. Circulation du virus dengue 2 dans plusieurs milieux boisés des savanes soudaniennes de la région de Bobo-Dioulasso (Burkina Faso). Cahier ORSTOM Série Entomol. Méd. et Parasitol. 1984, 22, 135–143. [Google Scholar]
- Robin, Y.; Cornet, M.; Heme, G.; Le Gonidec, G. Isolement du virus de la dengue au Senegal. Ann. Virol. (Institut Pasteur) 1980, 131, 149–154. [Google Scholar] [CrossRef]
- Saluzzo, J.F.; Cornet, M.; Adam, C.; Eyraud, M.; Digoutte, J.P. Dengue 2 in eastern Senegal: Serologic survey in simian and human populations. 1974–85. Bull. Soc. Pathol. Exot. Filiales 1986, 79, 313–322. [Google Scholar] [PubMed]
- Rudnick, A. Studies of the ecology of dengue in Malaysia: A preliminary report. J. Med. Entomol. 1965, 2, 203–208. [Google Scholar] [CrossRef] [PubMed]
- Yuwono, J.; Suharyono, W.; Koiman, I.; Tsuchiya, Y.; Tagaya, I. Seroepidemiological survey on dengue and Japanese encephalitis virus infections in Asian monkeys. Southeast Asian J. Trop. Med. Publ. Health 1984, 15, 194–200. [Google Scholar]
- Rosen, L. Experimental infection of new world monkeys with dengue and yellow fever viruses. Am. J. Trop. Med. Hyg. 1958, 7, 406–410. [Google Scholar] [CrossRef]
- Rosen, L. Observations on the epidemilogy of dengue in Panama. Am. J. Hyg. 1958, 68, 45–58. [Google Scholar] [PubMed]
- Roberts, D.R.; Peyton, E.L.; Pinheiro, F.P.; Balderrama, F.; Vargas, R. Associations of arbovirus vectors with gallery forests and domestic environments in southeastern Bolivia. Bull. Pan Am. Health Organ. 1984, 18, 337–350. [Google Scholar] [PubMed]
- Germain, M.; Sureau, P.; Herve, J.P.; Fabre, J.; Mouchet, J.; Robin, Y.; Geoffroy, B. Isolements du virus de la fievre jaune a partir d’ aedes du groupe A. africanus (theobald) en Republique Central Africaine. Importance des savanes humides et semi-humides en tant que zone d’emergence du virus amaril. Cah. ORSTOM. ser Ent. Med. et Parasitol. 1976, 14, 125–139. [Google Scholar]
- Rudnick, A. Ecology of dengue virus. Asian J. Infect. Dis. 1978, 2, 156–160. [Google Scholar]
- Mattingly, P.F. Genetical aspects of the Aedes aegypti problem. I. Taxonomy and bionomics. Ann. Trop. Med. Parasitol. 1957, 51, 392–408. [Google Scholar] [CrossRef]
- Gubler, D.J.; Rosen, L. Variation among geographic strains of Aedes albopictus in susceptibility to infection with dengue viruses. Am. J. Trop. Med. Hyg. 1976, 25, 318–325. [Google Scholar] [CrossRef]
- Metselaar, D.; Grainger, C.R.; Oei, K.G.; Reynolds, D.G.; Pudney, M.; Leake, C.J.; Tukei, P.M.; D’Offay, R.M.; Simpson, D.I. An outbreak of type 2 dengue fever in the Seychelles, probably transmitted by Aedes albopictus (skuse). Bull. World Health Organ. 1980, 58, 937–943. [Google Scholar] [PubMed]
- Christophers, S.R. Systematic. In Aedes aegypti, the Yellow Fever Mosquito: Its Life History, Bionomics and Structure; Cambridge University Press: London, UK, 1960; pp. 21–53. [Google Scholar]
- Edwards, F.W. Diptera.Family culicidae. In Genera Insectorum; Wystman, P., Ed.; Desmet-Verteneuil: Brussels, Belgium, 1932; Volume 194. [Google Scholar]
- Barraud, P.J. The distribution of ’stegomyia fasciata’ in India, with remarks on dengue and yellow fever. Indian J. Med. Res. 1928, 16, 377–398. [Google Scholar]
- Platt, K.B.; Linthicum, K.J.; Myint, K.S.; Innis, B.L.; Lerdthusnee, K.; Vaughn, D.W. Impact of dengue virus infection on feeding behavior of Aedes aegypti. Am. J. Trop. Med. Hyg. 1997, 57, 119–125. [Google Scholar] [CrossRef] [PubMed]
- Putnam, J.L.; Scott, T.W. Blood-feeding behavior of dengue-2 virus-infected Aedes aegypti. Am. J. Trop. Med. Hyg. 1995, 52, 225–227. [Google Scholar] [CrossRef]
- Harrington, L.C.; Edman, J.D.; Scott, T.W. Why do female Aedes aegypti (diptera: Culicidae) feed preferentially and frequently on human blood? J. Med. Entomol. 2001, 38, 411–422. [Google Scholar] [CrossRef]
- Gubler, D.J.; Nalim, S.; Tan, R.; Saipan, H.; Sulianti Saroso, J. Variation in susceptibility to oral infection with dengue viruses among geographic strains of Aedes aegypti. Am. J. Trop. Med. Hyg. 1979, 28, 1045–1052. [Google Scholar] [CrossRef]
- Libraty, D.H.; Endy, T.P.; Houng, H.S.; Green, S.; Kalayanarooj, S.; Suntayakorn, S.; Chansiriwongs, W.; Vaughn, D.W.; Nisalak, A.; Ennis, F.A.; et al. Differing influences of virus burden and immune activation on disease severity in secondary dengue-3 virus infections. J. Infect. Dis. 2002, 185, 1213–1221. [Google Scholar] [CrossRef]
- Khin, M.M.; Than, K.A. Transovarial transmission of dengue 2 virus by Aedes aegypti in nature. Am. J. Trop. Med. Hyg. 1983, 32, 590–594. [Google Scholar] [CrossRef]
- Rosen, L.; Shroyer, D.A.; Tesh, R.B.; Freier, J.E.; Lien, J.C. Transovarial transmission of dengue viruses by mosquitoes: Aedes albopictus and Aedes aegypti. Am. J. Trop. Med. Hyg. 1983, 32, 1108–1119. [Google Scholar] [CrossRef]
- Hull, B.; Tikasingh, E.; de Souza, M.; Martinez, R. Natural transovarial transmission of dengue 4 virus in Aedes aegypti in Trinidad. Am. J. Trop. Med. Hyg. 1984, 33, 1248–1250. [Google Scholar] [CrossRef]
- Joshi, V.; Singhi, M.; Chaudhary, R.C. Transovarial transmission of dengue 3 virus by Aedes aegypti. Trans. R. Soc. Trop. Med. Hyg. 1996, 90, 643–644. [Google Scholar] [CrossRef]
- Thavara, U.; Siriyasatien, P.; Tawatsin, A.; Asavadachanukorn, P.; Anantapreecha, S.; Wongwanich, R.; Mulla, M.S. Double infection of heteroserotypes of dengue viruses in field populations of Aedes aegypti and Aedes albopictus (diptera: Culicidae) and serological features of dengue viruses found in patients in southern Thailand. Southeast Asian J. Trop. Med. Publ. Health 2006, 37, 468–476. [Google Scholar]
- Thongrungkiat, S.; Maneekan, P.; Wasinpiyamongkol, L.; Prummongkol, S. Prospective field study of transovarial dengue-virus transmission by two different forms of Aedes aegypti in an urban area of Bangkok, Thailand. J. Vector Ecol. 2011, 36, 147–152. [Google Scholar] [CrossRef]
- Guedes, D.R.; Cordeiro, M.T.; Melo-Santos, M.A.; Magalhaes, T.; Marques, E.; Regis, L.; Furtado, A.F.; Ayres, C.F. Patient-based dengue virus surveillance in Aedes aegypti from Recife, Brazil. J. Vector Borne Dis. 2010, 47, 67–75. [Google Scholar]
- Zeidler, J.D.; Acosta, P.O.; Barreto, P.P.; Cordeiro Jda, S. Dengue virus in Aedes aegypti larvae and infestation dynamics in Roraima, Brazil. Rev. Saude Publ. 2008, 42, 986–991. [Google Scholar] [CrossRef]
- Arunachalam, N.; Tewari, S.C.; Thenmozhi, V.; Rajendran, R.; Paramasivan, R.; Manavalan, R.; Ayanar, K.; Tyagi, B.K. Natural vertical transmission of dengue viruses by Aedes aegypti in Chennai, Tamil Nadu, India. Ind. J. Med. Res. 2008, 127, 395–397. [Google Scholar]
- Gunther, J.; Martinez-Munoz, J.P.; Perez-Ishiwara, D.G.; Salas-Benito, J. Evidence of vertical transmission of dengue virus in two endemic localities in the state of Oaxaca, Mexico. Intervirology 2007, 50, 347–352. [Google Scholar] [CrossRef]
- Mitchell, C.J.; Miller, B.R. Vertical transmission of dengue viruses by strains of Aedes albopictus recently introduced into Brazil. J. Am. Mosq. Contl. Assoc. 1990, 6, 251–253. [Google Scholar]
- Rosen, L. Further observations on the mechanism of vertical transmission of flaviviruses by aedes mosquitoes. Am. J. Trop. Med. Hyg. 1988, 39, 123–126. [Google Scholar] [CrossRef]
- Shroyer, D.A. Vertical maintenance of dengue-1 virus in sequential generations of Aedes albopictus. J. Am. Mosq. Contl. Assoc. 1990, 6, 312–314. [Google Scholar]
- Freier, J.E.; Rosen, L. Vertical transmission of dengue viruses by Aedes mediovittatus. Am. J. Trop. Med. Hyg. 1988, 39, 218–222. [Google Scholar] [CrossRef] [PubMed]
- Freier, J.E.; Rosen, L. Vertical transmission of dengue viruses by mosquitoes of the Aedes scutellaris group. Am. J. Trop. Med. Hyg. 1987, 37, 640–647. [Google Scholar] [CrossRef] [PubMed]
- Goncalvez, A.P.; Escalante, A.A.; Pujol, F.H.; Ludert, J.E.; Tovar, D.; Salas, R.A.; Liprandi, F. Diversity and evolution of the envelope gene of dengue virus type 1. Virology 2002, 303, 110–119. [Google Scholar] [CrossRef] [PubMed]
- Wittke, V.; Robb, T.E.; Thu, H.M.; Nisalak, A.; Nimmannitya, S.; Kalayanrooj, S.; Vaughn, D.W.; Endy, T.P.; Holmes, E.C.; Aaskov, J.G. Extinction and rapid emergence of strains of dengue 3 virus during an interepidemic period. Virology 2002, 301, 148–156. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Mammen, M.P., Jr.; Chinnawirotpisan, P.; Klungthong, C.; Rodpradit, P.; Monkongdee, P.; Nimmannitya, S.; Kalayanarooj, S.; Holmes, E.C. Clade replacements in dengue virus serotypes 1 and 3 are associated with changing serotype prevalence. J. Virol. 2005, 79, 15123–15130. [Google Scholar] [CrossRef] [PubMed]
- Vezza, A.C.; Rosen, L.; Repik, P.; Dalrymple, J.; Bishop, D.H. Characterization of the viral RNA species of prototype dengue viruses. Am. J. Trop. Med. Hyg. 1980, 29, 643–652. [Google Scholar] [CrossRef] [PubMed]
- Repik, P.M.; Dalrymple, J.M.; Brandt, W.E.; McCown, J.M.; Russell, P.K. RNA fingerprinting as a method for distinguishing dengue 1 virus strains. Am. J. Trop. Med. Hyg. 1983, 32, 577–589. [Google Scholar] [CrossRef]
- Henchal, E.A.; Repik, P.M.; McCown, J.M.; Brandt, W.E. Identification of an antigenic and genetic variant of dengue-4 virus from the Caribbean. Am. J. Trop. Med. Hyg. 1986, 35, 393–400. [Google Scholar] [CrossRef] [PubMed]
- Kanakaratne, N.; Wahala, W.M.P.B.; Messer, W.B.; Tissera, H.A.; Shahani, A.; Abeysinghe, N.; de Silva, A.M.; Gunasekara, M.B. Severe dengue epidemics in Sri Lanka, 2003–2006. Emerg. Infect. Dis. 2009, 15, 192–199. [Google Scholar] [CrossRef]
- Mendez, J.A.; Usme-Ciro, J.A.; Domingo, C.; Rey, G.J.; Sanchez, J.A.; Tenorio, A.; Gallego-Gomez, J.C. Phylogenetic history demonstrates two different lineages of dengue type 1 virus in Colombia. Virol. J. 2010, 7, 226. [Google Scholar] [CrossRef]
- Teoh, B.T.; Sam, S.S.; Abd-Jamil, J.; AbuBakar, S. Isolation of ancestral sylvatic dengue virus type 1, Malaysia. Emerg. Infect. Dis. 2010, 16, 1783–1785. [Google Scholar] [CrossRef] [PubMed]
- Thu, H.M.; Lowry, K.; Myint, T.T.; Shwe, T.N.; Han, A.M.; Khin, K.K.; Thant, K.Z.; Thein, S.; Aaskov, J. Myanmar dengue outbreak associated with displacement of serotypes 2, 3, and 4 by dengue 1. Emerg. Infect. Dis. 2004, 10, 593–597. [Google Scholar] [CrossRef]
- Ong, S.H.; Yip, J.T.; Chen, Y.L.; Liu, W.; Harun, S.; Lystiyaningsih, E.; Heriyanto, B.; Beckett, C.G.; Mitchell, W.P.; Hibberd, M.L.; et al. Periodic re-emergence of endemic strains with strong epidemic potential—A proposed explanation for the 2004 Indonesian dengue epidemic. Infect. Genet. Evol. 2008, 8, 191–204. [Google Scholar] [CrossRef] [PubMed]
- Diaz, F.J.; Black, W.C.t.; Farfan-Ale, J.A.; Lorono-Pino, M.A.; Olson, K.E.; Beaty, B.J. Dengue virus circulation and evolution in Mexico: A phylogenetic perspective. Arch. Med. Res. 2006, 37, 760–773. [Google Scholar] [CrossRef] [PubMed]
- Zheng, K.; Zhou, H.Q.; Yan, J.; Ke, C.W.; Maeda, A.; Maeda, J.; Takashima, I.; Kurane, I.; Ma, H.; Xie, X.M. Molecular characterization of the E gene of dengue virus type 1 isolated in Guangdong province, China, in 2006. Epidemiol. Infect. 2009, 137, 73–78. [Google Scholar] [CrossRef] [PubMed]
- Cerdas-Quesada, C.A.; Retana-Salazar, A.P. Characterization and phylogenetic relationships of strains of dengue type 1 virus from Costa Rica. Rev. Biol. Trop. 2007, 55, 365–372. [Google Scholar] [PubMed]
- Myat Thu, H.; Lowry, K.; Jiang, L.; Hlaing, T.; Holmes, E.C.; Aaskov, J. Lineage extinction and replacement in dengue type 1 virus populations are due to stochastic events rather than to natural selection. Virology 2005, 336, 163–172. [Google Scholar] [CrossRef]
- Raghwani, J.; Rambaut, A.; Holmes, E.C.; Hang, V.T.; Hien, T.T.; Farrar, J.; Wills, B.; Lennon, N.J.; Birren, B.W.; Henn, M.R.; Simmons, C.P. Endemic dengue associated with the co-circulation of multiple viral lineages and localized density-dependent transmission. PLoS Pathog. 2011, 7, e1002064. [Google Scholar] [CrossRef]
- Imrie, A.; Roche, C.; Zhao, Z.; Bennett, S.; Laille, M.; Effler, P.; Cao-Lormeau, V.M. Homology of complete genome sequences for dengue virus type-1, from dengue-fever- and dengue-haemorrhagic-fever-associated epidemics in Hawaii and French Polynesia. Ann. Trop Med. Parasitol. 2010, 104, 225–235. [Google Scholar] [CrossRef]
- Osman, O.; Fong, M.Y.; Sekaran, S.D. Genetic characterization of dengue virus type 1 isolated in Brunei in 2005–2006. J. Gen. Virol. 2009, 90, 678–686. [Google Scholar] [CrossRef]
- Aviles, G.; Meissner, J.; Mantovani, R.; St Jeor, S. Complete coding sequences of dengue-1 viruses from Paraguay and Argentina. Virus Res. 2003, 98, 75–82. [Google Scholar] [CrossRef] [PubMed]
- Swofford, D.L. Paup*. Phylogenetic analysis using parsimony (* and othe methods). 4; Sinauer Associates: Sunderland, MA, USA, 2003. [Google Scholar]
- Posada, D.; Crandall, K.A. Modeltest: Testing the model of DNA substitution. Bioinformatics 1998, 14, 817–818. [Google Scholar] [CrossRef]
- A-Nuegoonpipat, A.; Berlioz-Arthaud, A.; Chow, V.; Endy, T.; Lowry, K.; Mai le, Q.; Ninh, T.U.; Pyke, A.; Reid, M.; Reynes, J.M.; et al. Sustained transmission of dengue virus type 1 in the Pacific due to repeated introductions of different asian strains. Virology 2004, 329, 505–512. [Google Scholar] [CrossRef] [PubMed]
- Nukui, Y.; Tajima, S.; Kotaki, A.; Ito, M.; Takasaki, T.; Koike, K.; Kurane, I. Novel dengue virus type 1 from travelers to Yap state, Micronesia. Emerg. Infect. Dis. 2006, 12, 343–346. [Google Scholar] [CrossRef]
- Shu, P.Y.; Su, C.L.; Liao, T.L.; Yang, C.F.; Chang, S.F.; Lin, C.C.; Chang, M.C.; Hu, H.C.; Huang, J.H. Molecular characterization of dengue viruses imported into Taiwan during 2003–2007: Geographic distribution and genotype shift. Am. J. Trop. Med. Hyg. 2009, 80, 1039–1046. [Google Scholar] [CrossRef] [PubMed]
- Yan, Y.S.; Hong, R.T.; Shen, X.N.; Weng, Y.W.; Cai, S.J.; Xu, B.H.; Li, S.Q.; He, J.X.; Xu, L.S.; Lin, Y.Q.; et al. Study on the epidemiology and etiologic agent of dengue fever outbreaks in Fuzhou in 2004. Zhonghua Liu Xing Bing Xue Za Zhi 2006, 27, 371–374. [Google Scholar]
- Zhang, F.C.; Chen, Y.Q.; Lu, Y.C.; Wang, J.; Chen, W.S.; Hong, W.X. Analysis on clinical and epidemiological characteristics of 1032 patients with dengue fever in Guangzhou. Zhonghua Liu Xing Bing Xue Za Zhi 2005, 26, 421–423. [Google Scholar]
- Xu, G.; Dong, H.; Shi, N.; Liu, S.; Zhou, A.; Cheng, Z.; Chen, G.; Liu, J.; Fang, T.; Zhang, H.; et al. An outbreak of dengue virus serotype 1 infection in Cixi, Ningbo, People’s Republic of China, 2004, associated with a traveler from Thailand and high density of Aedes albopictus. Am. J. Trop. Med. Hyg. 2007, 76, 1182–1188. [Google Scholar] [CrossRef]
- Imrie, A.; Zhao, Z.; Bennett, S.N.; Kitsutani, P.; Laille, M.; Effler, P. Molecular epidemiology of dengue in the Pacific: Introduction of two distinct strains of dengue virus type-1 into Hawaii. Ann. Trop. Med. Parasitol. 2006, 100, 327–336. [Google Scholar] [CrossRef]
- Morens, D.M.; Rigau-Perez, J.G.; Lopez-Correa, R.H.; Moore, C.G.; Ruiz-Tiben, E.E.; Sather, G.E.; Chiriboga, J.; Eliason, D.A.; Casta-Velez, A.; Woodall, J.P. Dengue in Puerto Rico, 1977: Public health response to characterize and control an epidemic of multiple serotypes. Am. J. Trop. Med. Hyg. 1986, 35, 197–211. [Google Scholar] [CrossRef]
- Rigau-Perez, J.G.; Gubler, D.J.; Vorndam, A.V.; Clark, G.G. Dengue surveillance—United States, 1986–1992. MMWR CDC Surveill. Sum. 1994, 43, 7–19. [Google Scholar]
- Gubler, D.J. Arboviruses as imported disease agents: The need for increased awareness. Arch. Virol. Suppl. 1996, 11, 21–32. [Google Scholar] [PubMed]
- Hafkin, B.; Kaplan, J.E.; Reed, C.; Elliott, L.B.; Fontaine, R.; Sather, G.E.; Kappus, K. Reintroduction of dengue fever into the continental United States. I. Dengue surveillance in Texas, 1980. Am. J. Trop. Med. Hyg. 1982, 31, 1222–1228. [Google Scholar] [CrossRef] [PubMed]
- Malison, M.D.; Waterman, S.H. Dengue fever in the United States. A report of a cluster of imported cases and review of the clinical, epidemiologic, and public health aspects of the disease. JAMA 1983, 249, 496–500. [Google Scholar] [CrossRef]
- Gubler, D.J. Dengue and dengue hemorrhagic fever. Clin. Microbiol. Rev. 1998, 11, 480–496. [Google Scholar] [CrossRef] [PubMed]
- Dantes, H.G.; Koopman, J.S.; Addy, C.L.; Zarate, M.L.; Marin, M.A.; Longini Junior, I.M.; Guttierez, E.S.; Rodriguez, V.A.; Garcia, L.G.; Mirelles, E.R. Dengue epidemics on the pacific coast of Mexico. Int. J. Epidemiol. 1988, 17, 178–186. [Google Scholar] [CrossRef] [PubMed]
- Herrera-Basto, E.; Prevots, D.R.; Zarate, M.L.; Silva, J.L.; Sepulveda-Amor, J. First reported outbreak of classical dengue fever at 1,700 meters above sea level in Guerrero state, Mexico, June 1988. Am. J. Trop. Med. Hyg. 1992, 46, 649–653. [Google Scholar] [CrossRef]
- Carvalho, S.E.; Martin, D.P.; Oliveira, L.M.; Ribeiro, B.M.; Nagata, T. Comparative analysis of American dengue virus type 1 full-genome sequences. Virus Gen. 2010, 40, 60–66. [Google Scholar] [CrossRef]
- Pires Neto, R.J.; Lima, D.M.; de Paula, S.O.; Lima, C.M.; Rocco, I.M.; Fonseca, B.A. Molecular epidemiology of type 1 and 2 dengue viruses in Brazil from 1988 to 2001. Braz. J. Med. Biol. Res. 2005, 38, 843–852. [Google Scholar] [CrossRef]
- Miagostovich, M.P.; Nogueira, R.M.; Cavalcanti, S.M.; Marzochi, K.B.; Schatzmayr, H.G. Dengue epidemic in the state of Rio de Janeiro, Brazil: Virological and epidemiological aspects. Rev. Inst. Med. Trop. Sao Paulo 1993, 35, 149–154. [Google Scholar] [CrossRef]
- Nogueira, R.M.; Schatzmayr, H.G.; Miagostovich, M.P.; Farias, M.F.; Farias Filho, J.D. Virological study of a dengue type 1 epidemic at Rio de Janeiro. Mem. Inst. Oswaldo Cruz 1988, 83, 219–225. [Google Scholar] [CrossRef]
- Aviles, G.; Paz, M.V.; Rangeon, G.; Ranaivoarisoa, M.Y.; Verzeri, N.; Roginski, S.; Baroni, P.; Enria, D. Laboratory surveillance of dengue in Argentina, 1995–2001. Emerg. Infect. Dis. 2003, 9, 738–742. [Google Scholar] [CrossRef] [PubMed]
- Guzman, M.G.; Kouri, G.; Valdes, L.; Bravo, J.; Alvarez, M.; Vazques, S.; Delgado, I.; Halstead, S.B. Epidemiologic studies on dengue in Santiago de Cuba, 1997. Am. J. Epidemiol. 2000, 152, 793–799; discussion 804. [Google Scholar] [CrossRef] [PubMed]
- Anonymous. Identification of human strains isolated in 1964, 1966, 1967 and 1968, as presumptive dengue virus. In 1968 Annual Report; University of Ibadan: Ibadan, Nigeria, 1968; pp. 122–123. [Google Scholar]
- Rico-Hesse, R.; Harrison, L.M.; Nisalak, A.; Vaughn, D.W.; Kalayanarooj, S.; Green, S.; Rothman, A.L.; Ennis, F.A. Molecular evolution of dengue type 2 virus in Thailand. Am. J. Trop. Med. Hyg. 1998, 58, 96–101. [Google Scholar] [CrossRef] [PubMed]
- Lewis, J.A.; Chang, G.J.; Lanciotti, R.S.; Kinney, R.M.; Mayer, L.W.; Trent, D.W. Phylogenetic relationships of dengue-2 viruses. Virology 1993, 197, 216–224. [Google Scholar] [CrossRef]
- Salda, L.T.; Parquet, M.D.; Matias, R.R.; Natividad, F.F.; Kobayashi, N.; Morita, K. Molecular epidemiology of dengue 2 viruses in The Philippines: Genotype shift and local evolution. Am. J. Trop. Med. Hyg. 2005, 73, 796–802. [Google Scholar] [CrossRef]
- Rabaa, M.A.; Ty Hang, V.T.; Wills, B.; Farrar, J.; Simmons, C.P.; Holmes, E.C. Phylogeography of recently emerged DENV-2 in southern Viet Nam. PLoS Negl. Trop. Dis. 2010, 4, e766. [Google Scholar] [CrossRef]
- Vu, T.T.; Holmes, E.C.; Duong, V.; Nguyen, T.Q.; Tran, T.H.; Quail, M.; Churcher, C.; Parkhill, J.; Cardosa, J.; Farrar, J.; et al. Emergence of the Asian 1 genotype of dengue virus serotype 2 in Viet Nam: In vivo fitness advantage and lineage replacement in south-east Asia. PLoS Negl. Trop. Dis. 2010, 4, e757. [Google Scholar]
- Huang, J.H.; Liao, T.L.; Chang, S.F.; Su, C.L.; Chien, L.J.; Kuo, Y.C.; Yang, C.F.; Lin, C.C.; Shu, P.Y. Laboratory-based dengue surveillance in Taiwan, 2005: A molecular epidemiologic study. Am. J. Trop. Med. Hyg. 2007, 77, 903–909. [Google Scholar] [CrossRef]
- Guzman, M.G.; Deubel, V.; Pelegrino, J.L.; Rosario, D.; Marrero, M.; Sariol, C.; Kouri, G. Partial nucleotide and amino acid sequences of the envelope and the envelope/nonstructural protein-1 gene junction of four dengue-2 virus strains isolated during the 1981 cuban epidemic. Am. J. Trop. Med. Hyg. 1995, 52, 241–246. [Google Scholar] [CrossRef]
- Rico-Hesse, R.; Harrison, L.M.; Salas, R.A.; Tovar, D.; Nisalak, A.; Ramos, C.; Boshell, J.; de Mesa, M.T.; Nogueira, R.M.; da Rosa, A.T. Origins of dengue type 2 viruses associated with increased pathogenicity in the Americas. Virology 1997, 230, 244–251. [Google Scholar] [CrossRef]
- Foster, J.E.; Bennett, S.N.; Carrington, C.V.; Vaughan, H.; McMillan, W.O. Phylogeography and molecular evolution of dengue 2 in the Caribbean basin, 1981–2000. Virology 2004, 324, 48–59. [Google Scholar] [CrossRef] [PubMed]
- Rico-Hesse, R. Microevolution and virulence of dengue viruses. Adv. Virus Res. 2003, 59, 315–341. [Google Scholar] [PubMed]
- Kouri, G.P.; Guzman, M.G.; Bravo, J.R.; Triana, C. Dengue haemorrhagic fever/dengue shock syndrome: Lessons from the cuban epidemic, 1981. Bull. World Health Org. 1989, 67, 375–380. [Google Scholar] [PubMed]
- Watts, D.M.; Porter, K.R.; Putvatana, P.; Vasquez, B.; Calampa, C.; Hayes, C.G.; Halstead, S.B. Failure of secondary infection with American genotype dengue 2 to cause dengue haemorrhagic fever. Lancet 1999, 354, 1431–1434. [Google Scholar] [CrossRef]
- Kumar, S.R.; Patil, J.A.; Cecilia, D.; Cherian, S.S.; Barde, P.V.; Walimbe, A.M.; Yadav, P.D.; Yergolkar, P.N.; Shah, P.S.; Padbidri, V.S.; et al. Evolution, dispersal and replacement of american genotype dengue type 2 viruses in India (1956–2005): Selection pressure and molecular clock analyses. J. Gen. Virol. 2010, 91, 707–720. [Google Scholar] [CrossRef]
- Dorji, T.; Yoon, I.K.; Holmes, E.C.; Wangchuk, S.; Tobgay, T.; Nisalak, A.; Chinnawirotpisan, P.; Sangkachantaranon, K.; Gibbons, R.V.; Jarman, R.G. Diversity and origin of dengue virus serotypes 1, 2, and 3, Bhutan. Emerg. Infect. Dis. 2009, 15, 1630–1632. [Google Scholar] [CrossRef]
- Zaki, A.; Perera, D.; Jahan, S.S.; Cardosa, M.J. Phylogeny of dengue viruses circulating in Jeddah, Saudi Arabia: 1994 to 2006. Trop. Med. Int. Health 2008, 13, 584–592. [Google Scholar] [CrossRef]
- Kanesa-thasan, N.; Chang, G.J.; Smoak, B.L.; Magill, A.; Burrous, M.J.; Hoke, C.H., Jr. Molecular and epidemiologic analysis of dengue virus isolates from Somalia. Emerg. Infect. Dis. 1998, 4, 299–303. [Google Scholar] [CrossRef]
- Huhtamo, E.; Uzcategui, N.Y.; Siikamaki, H.; Saarinen, A.; Piiparinen, H.; Vaheri, A.; Vapalahti, O. Molecular epidemiology of dengue virus strains from Finnish travelers. Emerg. Infect. Dis. 2008, 14, 80–83. [Google Scholar] [CrossRef]
- Holmes, E.C.; Tio, P.H.; Perera, D.; Muhi, J.; Cardosa, J. Importation and co-circulation of multiple serotypes of dengue virus in Sarawak, Malaysia. Virus Res. 2009, 143, 1–5. [Google Scholar] [CrossRef]
- Lee, K.S.; Lai, Y.L.; Lo, S.; Barkham, T.; Aw, P.; Ooi, P.L.; Tai, J.C.; Hibberd, M.; Johansson, P.; Khoo, S.P.; et al. Dengue virus surveillance for early warning, Singapore. Emerg. Infect. Dis. 2010, 16, 847–849. [Google Scholar] [CrossRef]
- Tung, Y.C.; Lin, K.H.; Chiang, H.C.; Ke, L.Y.; Chen, Y.H.; Ke, G.M.; Chen, T.C.; Chou, L.C.; Lu, P.L. Molecular epidemiology of dengue virus serotype 2 in the Taiwan 2002 outbreak with envelope gene and nonstructural protein 1 gene analysis. Kaohsiung J. Med. Sci. 2008, 24, 398–407. [Google Scholar] [CrossRef]
- Chen, H.L.; Lin, S.R.; Liu, H.F.; King, C.C.; Hsieh, S.C.; Wang, W.K. Evolution of dengue virus type 2 during two consecutive outbreaks with an increase in severity in southern Taiwan in 2001–2002. Am. J. Trop. Med. Hyg. 2008, 79, 495–505. [Google Scholar] [CrossRef] [PubMed]
- Singh, U.B.; Maitra, A.; Broor, S.; Rai, A.; Pasha, S.T.; Seth, P. Partial nucleotide sequencing and molecular evolution of epidemic causing dengue 2 strains. J. Infect. Dis. 1999, 180, 959–965. [Google Scholar] [CrossRef] [PubMed]
- Vasilakis, N.; Shell, E.J.; Fokam, E.B.; Mason, P.W.; Hanley, K.A.; Estes, D.M.; Weaver, S.C. Potential of ancestral sylvatic dengue-2 viruses to re-emerge. Virology 2007, 358, 402–412. [Google Scholar] [CrossRef] [PubMed]
- Leitmeyer, K.C.; Vaughn, D.W.; Watts, D.M.; Salas, R.; Villalobos, I.; de, C.; Ramos, C.; Rico-Hesse, R. Dengue virus structural differences that correlate with pathogenesis. J. Virol. 1999, 73, 4738–4747. [Google Scholar] [CrossRef]
- Steel, A.; Gubler, D.J.; Bennett, S.N. Natural attenuation of dengue virus type-2 after a series of island outbreaks: A retrospective phylogenetic study of events in the South Pacific three decades ago. Virology 2010, 405, 505–512. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Correa, R.H.; Cline, B.L.; Ramirez-Ronda, C.; Bermudez, R.; Sather, G.E.; Kuno, G. Dengue fever with hemorrhagic manifestations: A report of three cases from Puerto Rico. Am. J. Trop. Med. Hyg. 1978, 27, 1216–1224. [Google Scholar] [CrossRef]
- Moreau, J.P.; Rosen, L.; Saugrain, J.; Lagraulet, J. An epidemic of dengue on Tahiti associated with hemorrhagic manifestations. Am. J. Trop. Med. Hyg. 1973, 22, 237–241. [Google Scholar] [CrossRef]
- Loison, G.; Rosen, L.; Papillaud, J.; Tomasini, J.; Vaujany, J.; Chanalet, G. La dengue en Nouvelle-Caledonie (1971–1972). Bull. Soc. Path. Exot. 1973, 66, 511–519. [Google Scholar] [CrossRef]
- Barnes, W.J.; Rosen, L. Fatal hemorrhagic disease and shock associated with primary dengue infection on a pacific island. Am. J. Trop. Med. Hyg. 1974, 23, 495–506. [Google Scholar] [CrossRef]
- Carey, D.E.; Causey, O.R.; Reddy, S.; Cooke, A.R. Dengue viruses from febrile patients in Nigeria, 1964–68. Lancet 1971, 1, 105–106. [Google Scholar] [CrossRef] [PubMed]
- Cardosa, J.; Ooi, M.H.; Tio, P.H.; Perera, D.; Holmes, E.C.; Bibi, K.; Abdul Manap, Z. Dengue virus serotype 2 from a sylvatic lineage isolated from a patient with dengue hemorrhagic fever. PLoS Negl. Trop. Dis. 2009, 3, e423. [Google Scholar] [CrossRef] [PubMed]
- Zeller, H.G.; Traore-Lamizana, M.; Monlun, E.; Hervy, J.P.; Mondo, M.; Digoutte, J.P. Dengue-2 virus isolation from humans during an epizootic in southeastern Senegal in November, 1990. Res. Virol. 1992, 143, 101–102. [Google Scholar] [CrossRef]
- Monlun, E.; Zeller, H.; Traore-Lamizana, M.; Hervy, J.P.; Adam, F.; Mondo, M.; Digoutte, J.P. Caracteres cliniques et epidemiologiques de la dengue 2 au Senegal. Med. Mal. Infect. 1992, 22, 718–721. [Google Scholar] [CrossRef]
- Saluzzo, J.F.; Cornet, M.; Castagnet, P.; Rey, C.; Digoutte, J.P. Isolation of dengue 2 and dengue 4 viruses from patients in Senegal. Trans. R. Soc. Trop. Med. Hyg. 1986, 80, 5. [Google Scholar] [CrossRef] [PubMed]
- Franco, L.; Palacios, G.; Martinez, J.A.; Vazquez, A.; Savji, N.; Ory, F.D.; Sanchez-Seco, M.P.; Martin, D.; Lipkin, W.I.; Tenorio, A. First report of sylvatic DENV-2-associated dengue hemorrhagic fever in West Africa. PLoS Negl. Trop. Dis. 2011, 5, e1251. [Google Scholar] [CrossRef]
- Hervy, J.P.; Legros, F.; Roche, J.C.; Monteny, N.; Diaco, B. Circulation du dengue 2 dans plusieurs milieux boises des savanes soudaniennes de la region de Bobo-Dioulasso (Burkina Faso). Considerations entomologiques et epidemiologiques. Cah. ORSTOM. ser Ent. Med. et Parasitol. 1984, 22, 135–143. [Google Scholar]
- Vasilakis, N.; Holmes, E.C.; Fokam, E.B.; Faye, O.; Diallo, M.; Sall, A.A.; Weaver, S.C. Evolutionary processes among sylvatic dengue-2 viruses. J. Virol. 2007, 81, 9591–9595. [Google Scholar] [CrossRef]
- Lanciotti, R.S.; Lewis, J.G.; Gubler, D.J.; Trent, D.W. Molecular evolution and epidemiology of dengue-3 viruses. J. Gen. Virol. 1994, 75, 65–75. [Google Scholar] [CrossRef]
- Kochel, T.; Aguilar, P.; Felices, V.; Comach, G.; Cruz, C.; Alava, A.; Vargas, J.; Olson, J.; Blair, P. Molecular epidemiology of dengue virus type 3 in northern South America: 2000–2005. Infect. Genet. Evol. 2008, 8, 682–688. [Google Scholar] [CrossRef]
- Podder, G.; Breiman, R.F.; Azim, T.; Thu, H.M.; Velathanthiri, N.; Mai le, Q.; Lowry, K.; Aaskov, J.G. Origin of dengue type 3 viruses associated with the dengue outbreak in Dhaka, Bangladesh, in 2000 and 2001. Am. J. Trop. Med. Hyg. 2006, 74, 263–265. [Google Scholar] [CrossRef] [PubMed]
- Uzcategui, N.Y.; Comach, G.; Camacho, D.; Salcedo, M.; Cabello de Quintana, M.; Jimenez, M.; Sierra, G.; Cuello de Uzcategui, R.; James, W.S.; Turner, S.; et al. Molecular epidemiology of dengue virus type 3 in Venezuela. J. Gen. Virol. 2003, 84, 1569–1575. [Google Scholar] [CrossRef] [PubMed]
- Aquino, V.H.; Anatriello, E.; Goncalves, P.F.; EV, D.A.S.; Vasconcelos, P.F.; Vieira, D.S.; Batista, W.C.; Bobadilla, M.L.; Vazquez, C.; Moran, M.; Figueiredo, L.T. Molecular epidemiology of dengue type 3 virus in Brazil and Paraguay, 2002–2004. Am. J. Trop. Med. Hyg. 2006, 75, 710–715. [Google Scholar] [CrossRef] [PubMed]
- Ramirez, A.; Fajardo, A.; Moros, Z.; Gerder, M.; Caraballo, G.; Camacho, D.; Comach, G.; Alarcon, V.; Zambrano, J.; Hernandez, R.; et al. Evolution of dengue virus type 3 genotype III in Venezuela: Diversification, rates and population dynamics. Virol. J. 2010, 7, 329. [Google Scholar] [CrossRef]
- Moi, M.L.; Takasaki, T.; Kotaki, A.; Tajima, S.; Lim, C.K.; Sakamoto, M.; Iwagoe, H.; Kobayashi, K.; Kurane, I. Importation of dengue virus type 3 to Japan from Tanzania and Cote d’Ivoire. Emerg. Infect. Dis. 2010, 16, 1770–1772. [Google Scholar] [CrossRef]
- Ospina, M.C.; Diaz, F.J.; Osorio, J.E. Prolonged co-circulation of two distinct dengue virus type 3 lineages in the hyperendemic area of Medellin, Colombia. Am. J. Trop. Med. Hyg. 2010, 83, 672–678. [Google Scholar] [CrossRef]
- Amarilla, A.A.; de Almeida, F.T.; Jorge, D.M.; Alfonso, H.L.; de Castro-Jorge, L.A.; Nogueira, N.A.; Figueiredo, L.T.; Aquino, V.H. Genetic diversity of the E protein of dengue type 3 virus. Virol. J. 2009, 6, 113. [Google Scholar] [CrossRef]
- Villabona-Arenas, C.J.; Miranda-Esquivel, D.R.; Jimenez, R.E. Phylogeny of dengue virus type 3 circulating in Colombia between 2001 and 2007. Trop. Med. Int. Health 2009, 14, 1241–1250. [Google Scholar] [CrossRef]
- de Mora, D.; Andrea, L.D.; Alvarez, M.; Regato, M.; Fajardo, A.; Recarey, R.; Colina, R.; Khan, B.; Cristina, J. Evidence of diversification of dengue virus type 3 genotype III in the South American region. Arch. Virol. 2009, 154, 699–707. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, N.; Thayan, R.; Sugimoto, C.; Oda, K.; Saat, Z.; Vijayamalar, B.; Sinniah, M.; Igarashi, A. Type-3 dengue viruses responsible for the dengue epidemic in Malaysia during 1993–1994. Am. J. Trop. Med. Hyg. 1999, 60, 904–909. [Google Scholar] [CrossRef] [PubMed]
- Chow, V.T.; Seah, C.L.; Chan, Y.C. Comparative analysis of NS3 sequences of temporally separated dengue 3 virus strains isolated from Southeast Asia. Intervirology 1994, 37, 252–258. [Google Scholar] [CrossRef] [PubMed]
- Dash, P.K.; Parida, M.M.; Saxena, P.; Abhyankar, A.; Singh, C.P.; Tewari, K.N.; Jana, A.M.; Sekhar, K.; Rao, P.V. Reemergence of dengue virus type-3 (subtype-III) in India: Implications for increased incidence of dhf & dss. Virol. J. 2006, 3, 55. [Google Scholar]
- Usuku, S.; Castillo, L.; Sugimoto, C.; Noguchi, Y.; Yogo, Y.; Kobayashi, N. Phylogenetic analysis of dengue-3 viruses prevalent in Guatemala during 1996–1998. Arch. Virol. 2001, 146, 1381–1390. [Google Scholar] [CrossRef]
- Chao, D.Y.; King, C.C.; Wang, W.K.; Chen, W.J.; Wu, H.L.; Chang, G.J. Strategically examining the full-genome of dengue virus type 3 in clinical isolates reveals its mutation spectra. Virol. J. 2005, 2, 72. [Google Scholar] [CrossRef]
- Sharma, S.; Dash, P.K.; Agarwal, S.K.; Shukla, J.; Parida, M.M.; Rao, P.V. Comparative complete genome analysis of dengue virus type 3 circulating in India between 2003–2008. J. Gen. Virol. 2011, in press. [Google Scholar] [CrossRef]
- King, C.C.; Chao, D.Y.; Chien, L.J.; Chang, G.J.; Lin, T.H.; Wu, Y.C.; Huang, J.H. Comparative analysis of full genomic sequences among different genotypes of dengue virus type 3. Virol. J. 2008, 5, 63. [Google Scholar] [CrossRef]
- Nogueira, M.B.; Stella, V.; Bordignon, J.; Batista, W.C.; Borba, L.; Silva, L.H.; Hoffmann, F.G.; Probst, C.M.; Santos, C.N. Evidence for the co-circulation of dengue virus type 3 genotypes III and V in the northern region of Brazil during the 2002–2004 epidemics. Mem. Inst. Oswaldo Cruz 2008, 103, 483–488. [Google Scholar] [CrossRef]
- Araujo, J.M.; Bello, G.; Schatzmayr, H.G.; Santos, F.B.; Nogueira, R.M. Dengue virus type 3 in Brazil: A phylogenetic perspective. Mem. Inst. Oswaldo Cruz 2009, 104, 526–529. [Google Scholar] [CrossRef]
- Chungue, E.; Deubel, V.; Cassar, O.; Laille, M.; Martin, P.M. Molecular epidemiology of dengue 3 viruses and genetic relatedness among dengue 3 strains isolated from patients with mild or severe form of dengue fever in French Polynesia. J. Gen. Virol. 1993, 74, 2765–2770. [Google Scholar] [CrossRef] [PubMed]
- Raekiansyah, M.; Pramesyanti, A.; Bela, B.; Kosasih, H.; Ma’roef, C.N.; Tobing, S.Y.; Rudiman, P.I.; Alisjahbana, B.; Endi, T.P.; Green, S.; et al. Genetic variations and relationship among dengue virus type 3 strains isolated from patients with mild or severe form of dengue disease in Indonesia and Thailand. Southeast Asian J. Trop. Med. Publ. Health 2005, 36, 1187–1197. [Google Scholar]
- Corwin, A.L.; Larasati, R.P.; Bangs, M.J.; Wuryadi, S.; Arjoso, S.; Sukri, N.; Listyaningsih, E.; Hartati, S.; Namursa, R.; Anwar, Z.; et al. Epidemic dengue transmission in southern Sumatra, Indonesia. Trans. R. Soc. Trop. Med. Hyg. 2001, 95, 257–265. [Google Scholar] [CrossRef] [PubMed]
- Gubler, D.J.; Suharyono, W.; Lubis, I.; Eram, S.; Gunarso, S. Epidemic dengue 3 in Central Java, associated with low viremia in man. Am. J. Trop. Med. Hyg. 1981, 30, 1094–1099. [Google Scholar] [CrossRef] [PubMed]
- Gubler, D.J.; Suharyono, W.; Tan, R.; Abidin, M.; Sie, A. Viraemia in patients with naturally acquired dengue infection. Bull. World Health Organ. 1981, 59, 623–630. [Google Scholar] [PubMed]
- Kitchener, S.; Leggat, P.A.; Brennan, L.; McCall, B. Importation of dengue by soldiers returning from East Timor to North Queensland, Australia. J. Travel Med. 2002, 9, 180–183. [Google Scholar] [CrossRef]
- Ito, M.; Takasaki, T.; Kotaki, A.; Tajima, S.; Yuwono, D.; Rimal, H.S.; dos Santos, F.; de Jesus, M.D.; Lina, B.B.; Tsuda, Y.; et al. Molecular and virological analyses of dengue virus responsible for dengue outbreak in East Timor in 2005. Jpn. J. Infect. Dis. 2010, 63, 181–184. [Google Scholar] [CrossRef]
- Kalayanarooj, S.; Rimal, H.S.; Andjaparidze, A.; Vatcharasaevee, V.; Nisalak, A.; Jarman, R.G.; Chinnawirotpisan, P.; Mammen, M.P.; Holmes, E.C.; Gibbons, R.V. Clinical intervention and molecular characteristics of a dengue hemorrhagic fever outbreak in Timor Leste, 2005. Am. J. Trop. Med. Hyg. 2007, 77, 534–537. [Google Scholar] [CrossRef] [PubMed]
- Chow, V.T.; Seah, C.L.; Chan, Y.C. Use of NS3 consensus primers for the polymerase chain reaction amplification and sequencing of dengue viruses and other flaviviruses. Arch. Virol. 1993, 133, 157–170. [Google Scholar] [CrossRef]
- Chungue, E.; Spiegel, A.; Roux, J.; Laudon, F.; Cardines, R. Dengue-3 in French Polynesia: Preliminary data. Med. J. Aust. 1990, 152, 557–558. [Google Scholar] [CrossRef]
- Barcelos Figueiredo, L.; Batista Cecilio, A.; Portela Ferreira, G.; Paiva Drumond, B.; Germano de Oliveira, J.; Bonjardim, C.A.; Peregrino Ferreira, P.C.; Kroon, E.G. Dengue virus 3 genotype 1 associated with dengue fever and dengue hemorrhagic fever, Brazil. Emerg. Infect. Dis. 2008, 14, 314–316. [Google Scholar] [CrossRef] [PubMed]
- Islam, M.A.; Ahmed, M.U.; Begum, N.; Chowdhury, N.A.; Khan, A.H.; Parquet Mdel, C.; Bipolo, S.; Inoue, S.; Hasebe, F.; Suzuki, Y.; et al. Molecular characterization and clinical evaluation of dengue outbreak in 2002 in Bangladesh. Jpn. J. Infect. Dis. 2006, 59, 85–91. [Google Scholar] [PubMed]
- Aziz, M.M.; Hasan, K.N.; Hasanat, M.A.; Siddiqui, M.A.; Salimullah, M.; Chowdhury, A.K.; Ahmed, M.; Alam, M.N.; Hassan, M.S. Predominance of the DEN-3 genotype during the recent dengue outbreak in Bangladesh. Southeast Asian J. Trop. Med. Publ. Health 2002, 33, 42–48. [Google Scholar]
- Ito, M.; Yamada, K.; Takasaki, T.; Pandey, B.; Nerome, R.; Tajima, S.; Morita, K.; Kurane, I. Phylogenetic analysis of dengue viruses isolated from imported dengue patients: Possible aid for determining the countries where infections occurred. J. Travel Med. 2007, 14, 233–244. [Google Scholar] [CrossRef] [PubMed]
- Vainio, K.; Noraas, S.; Holmberg, M.; Fremstad, H.; Wahlstrom, M.; Anestad, G.; Dudman, S. Fatal and mild primary dengue virus infections imported to Norway from Africa and south-east Asia, 2008–2010. Euro Surveill. 2010, 15, pii=19666. [Google Scholar] [CrossRef] [PubMed]
- Gubler, D.J.; Sather, G.E.; Kuno, G.; Cabral, J.R. Dengue 3 virus transmission in Africa. Am. J. Trop. Med. Hyg. 1986, 35, 1280–1284. [Google Scholar] [CrossRef]
- CDC. Dengue type 3 infection—Nicaragua and Panama, October-November 1994. JAMA 1995, 273, 840–841. [Google Scholar] [CrossRef]
- Guzman, M.G.; Vazquez, S.; Martinez, E.; Alvarez, M.; Rodriguez, R.; Kouri, G.; de los Reyes, J.; Acevedo, F. Dengue in Nicaragua, 1994: Reintroduction of serotype 3 in the Americas. Bol. Oficina Sanit. Panam. 1996, 121, 102–110. [Google Scholar] [CrossRef]
- Briseno-Garcia, B.; Gomez-Dantes, H.; Argott-Ramirez, E.; Montesano, R.; Vazquez-Martinez, A.L.; Ibanez-Bernal, S.; Madrigal-Ayala, G.; Ruiz-Matus, C.; Flisser, A.; Tapia-Conyer, R. Potential risk for dengue hemorrhagic fever: The isolation of serotype dengue-3 in Mexico. Emerg. Infect. Dis. 1996, 2, 133–135. [Google Scholar] [CrossRef]
- Anonymous. Isolation of dengue type 3 virus prompts concern and action. Bull. Pan Am. Health Organ. 1995, 29, 184–185. [Google Scholar]
- Harris, E.; Sandoval, E.; Xet-Mull, A.M.; Johnson, M.; Riley, L.W. Rapid subtyping of dengue viruses by restriction site-specific (RSS)-PCR. Virology 1999, 253, 86–95. [Google Scholar] [CrossRef] [PubMed]
- Peyrefitte, C.N.; Couissinier-Paris, P.; Mercier-Perennec, V.; Bessaud, M.; Martial, J.; Kenane, N.; Durand, J.P.; Tolou, H.J. Genetic characterization of newly reintroduced dengue virus type 3 in Martinique (French West Indies). J. Clin. Microbiol. 2003, 41, 5195–5198. [Google Scholar] [CrossRef]
- Rigau-Perez, J.G.; Ayala-Lopez, A.; Garcia-Rivera, E.J.; Hudson, S.M.; Vorndam, V.; Reiter, P.; Cano, M.P.; Clark, G.G. The reappearance of dengue-3 and a subsequent dengue-4 and dengue-1 epidemic in Puerto Rico in 1998. Am. J. Trop. Med. Hyg. 2002, 67, 355–362. [Google Scholar] [CrossRef] [PubMed]
- CDC. Dengue outbreak associated with multiple serotypes—Puerto Rico, 1998. MMWR 1998, 47, 952–956. [Google Scholar]
- Peyrefitte, C.N.; Pastorino, B.A.; Bessaud, M.; Gravier, P.; Tock, F.; Couissinier-Paris, P.; Martial, J.; Huc-Anais, P.; Cesaire, R.; Grandadam, M.; et al. Dengue type 3 virus, Saint Martin, 2003–2004. Emerg. Infect. Dis. 2005, 11, 757–761. [Google Scholar] [CrossRef]
- Rodriguez-Roche, R.; Alvarez, M.; Holmes, E.C.; Bernardo, L.; Kouri, G.; Gould, E.A.; Halstead, S.; Guzman, M.G. Dengue virus type 3, Cuba, 2000–2002. Emerg. Infect. Dis. 2005, 11, 773–774. [Google Scholar] [CrossRef]
- Nogueira, R.M.; Miagostovich, M.P.; de Filippis, A.M.; Pereira, M.A.; Schatzmayr, H.G. Dengue virus type 3 in Rio de Janeiro, Brazil. Mem. Inst. Oswaldo Cruz 2001, 96, 925–926. [Google Scholar] [CrossRef]
- Ocazionez, R.E.; Cortes, F.M.; Villar, L.A.; Gomez, S.Y. Temporal distribution of dengue virus serotypes in Colombian endemic area and dengue incidence. Re-introduction of dengue-3 associated to mild febrile illness and primary infection. Mem. Inst. Oswaldo Cruz 2006, 101, 725–731. [Google Scholar] [CrossRef]
- Barrero, P.R.; Mistchenko, A.S. Genetic analysis of dengue virus type 3 isolated in Buenos Aires, Argentina. Virus Res. 2008, 135, 83–88. [Google Scholar] [CrossRef]
- Isturiz, R.E.; Gubler, D.J.; Brea del Castillo, J. Dengue and dengue hemorrhagic fever in Latin America and the Caribbean. Infect. Dis. Clin. North Am. 2000, 14, 121–140, ix. [Google Scholar] [CrossRef]
- Sun, J.; Lin, J.; Yan, J.; Fan, W.; Lu, L.; Lv, H.; Hou, J.; Ling, F.; Fu, T.; Chen, Z.; et al. Dengue virus serotype 3 subtype III, Zhejiang province, China. Emerg. Infect. Dis. 2011, 17, 321–323. [Google Scholar] [CrossRef]
- Neff, J.M.; Morris, L.; Gonzalez-Alcover, R.; Coleman, P.H.; Lyss, S.B.; Negron, H. Dengue fever in a Puerto Rican community. Am. J. Epidemiol. 1967, 86, 162–184. [Google Scholar] [CrossRef] [PubMed]
- Laigret, J.; Rosen, L.; Scholammer, G. On an epidemic of dengue occurring in Tahiti in 1964. Relations to the “Hemorrhagic fevers” Of Southeast Asia. Bull. Soc. Pathol. Exot. Filiales. 1967, 60, 339–353. [Google Scholar] [PubMed]
- Russell, P.K.; McCown, J.M. Comparison of dengue-2 and dengue-3 virus strains by neutralization tests and identification of a subtype of dengue-3. Am. J. Trop. Med. Hyg. 1972, 21, 97–99. [Google Scholar] [CrossRef] [PubMed]
- Saugrain, J.; Rosen, L.; Outin-Fabre, D.; Moreau, J.P. A recent epidemic due to arbovirus infections of the dengue type in Tahiti. Comparative study of the 1964 epidemic. Bull. Soc. Pathol. Exot. Filiales. 1970, 63, 636–642. [Google Scholar] [PubMed]
- Hammon, W.M.; Rudnick, A.; Sather, G. Viruses associated with epidemic hemorrhagic fevers of the Philippines and Thailand. Science 1960, 131, 1102–1103. [Google Scholar] [CrossRef]
- Di, B.; Bai, Z.J.; Wang, Y.L.; Luo, L.; Chen, Y.; Jiang, L.Y.; Yang, Z.C.; Wang, M. Molecular epidemiologic analysis on new emerged type 3 dengue virus in Guangzhou in 2009. Zhonghua Liu Xing Bing Xue Za Zhi 2010, 31, 804–807. [Google Scholar]
- Jenkins, G.M.; Rambaut, A.; Pybus, O.G.; Holmes, E.C. Rates of molecular evolution in RNA viruses: A quantitative phylogenetic analysis. J. Mol. Evol. 2002, 54, 156–165. [Google Scholar] [CrossRef]
- Klungthong, C.; Zhang, C.; Mammen, M.P., Jr.; Ubol, S.; Holmes, E.C. The molecular epidemiology of dengue virus serotype 4 in bangkok, Thailand. Virology 2004, 329, 168–179. [Google Scholar] [CrossRef] [PubMed]
- Lanciotti, R.S.; Gubler, D.J.; Trent, D.W. Molecular evolution and phylogeny of dengue-4 viruses. J. Gen. Virol. 1997, 78, 2279–2284. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Mammen, M.P., Jr.; Chinnawirotpisan, P.; Klungthong, C.; Rodpradit, P.; Nisalak, A.; Vaughn, D.W.; Nimmannitya, S.; Kalayanarooj, S.; Holmes, E.C. Structure and age of genetic diversity of dengue virus type 2 in Thailand. J. Gen. Virol. 2006, 87, 873–883. [Google Scholar] [CrossRef] [PubMed]
- Rudnick, A.; Lim, T.W. Dengue fever studies in Malaysia. Inst. Med. Res. Malaysia Bulletin 1986, 23, 51–152. [Google Scholar]
- Gubler, D. Personal communication. Duke/NUS, Singapore. 2011. [Google Scholar]
- AbuBakar, S.; Wong, P.F.; Chan, Y.F. Emergence of dengue virus type 4 genotype IIa in Malaysia. J. Gen. Virol. 2002, 83, 2437–2442. [Google Scholar] [CrossRef] [PubMed]
- Dash, P.K.; Sharma, S.; Srivastava, A.; Santhosh, S.R.; Parida, M.M.; Neeraja, M.; Subbalaxmi, M.V.; Lakshmi, V.; Rao, P.V. Emergence of dengue virus type 4 (genotype I) in India. Epidemiol. Infect. 2011, 139, 857–861. [Google Scholar] [CrossRef]
- Cecilia, D.; Kakade, M.B.; Bhagat, A.B.; Vallentyne, J.; Singh, A.; Patil, J.A.; Todkar, S.M.; Varghese, S.B.; Shah, P.S. Detection of dengue-4 virus in pune, western India after an absence of 30 years—Its association with two severe cases. Virol. J. 2011, 8, 46. [Google Scholar] [CrossRef]
- Figueiredo, R.M.; Naveca, F.G.; Bastos, M.S.; Melo, M.N.; Viana, S.S.; Farias, I.P. Dengue virus type 4, Manaus Brazil. Emerg. Infect. Dis. 2008, 14, 667–669. [Google Scholar] [CrossRef]
- Melo, F.L.; Romano, C.M.; Zanotto, P.M. Introduction of dengue virus 4 (DENV-4) genotype I into Brazil from Asia? PLoS Negl. Trop. Dis. 2009, 3, e390. [Google Scholar]
- Fernandez, J.; Vera, L.; Tognarelli, J.; Fasce, R.; Araya, P.; Villagra, E.; Roos, O.; Mora, J. Detection of dengue virus type 4 in Easter island, Chile. Arch. Virol. 2011, in press. [Google Scholar] [CrossRef]
- Foster, J.E.; Bennett, S.N.; Vaughan, H.; Vorndam, V.; McMillan, W.O.; Carrington, C.V. Molecular evolution and phylogeny of dengue type 4 virus in the Caribbean. Virology 2003, 306, 126–134. [Google Scholar] [CrossRef]
- Carrington, C.V.; Foster, J.E.; Pybus, O.G.; Bennett, S.N.; Holmes, E.C. Invasion and maintenance of dengue virus type 2 and type 4 in the Americas. J. Virol. 2005, 79, 14680–14687. [Google Scholar] [CrossRef]
- CDC. Dengue type 4 infections in U.S. travelers to the Caribbean. MMWR 1981, 30, 249–250. [Google Scholar]
- Bennett, S.N.; Drummond, A.J.; Kapan, D.D.; Suchard, M.A.; Munoz-Jordan, J.L.; Pybus, O.G.; Holmes, E.C.; Gubler, D.J. Epidemic dynamics revealed in dengue evolution. Mol. Biol. Evol. 2010, 27, 811–818. [Google Scholar] [CrossRef] [PubMed]
- Dussart, P.; Lavergne, A.; Lagathu, G.; Lacoste, V.; Martial, J.; Morvan, J.; Cesaire, R. Reemergence of dengue virus type 4, French Antilles and French Guiana, 2004–2005. Emerg. Infect. Dis. 2006, 12, 1748–1751. [Google Scholar] [CrossRef] [PubMed]
- Forshey, B.M.; Morrison, A.C.; Cruz, C.; Rocha, C.; Vilcarromero, S.; Guevara, C.; Camacho, D.E.; Alava, A.; Madrid, C.; Beingolea, L.; Suarez, V.; Comach, G.; Kochel, T.J. Dengue virus serotype 4, northeastern Peru, 2008. Emerg. Infect. Dis. 2009, 15, 1815–1818. [Google Scholar] [CrossRef] [PubMed]
- Bennett, S.N.; Holmes, E.C.; Chirivella, M.; Rodriguez, D.M.; Beltran, M.; Vorndam, V.; Gubler, D.J.; McMillan, W.O. Selection-driven evolution of emergent dengue virus. Mol. Biol. Evol. 2003, 20, 1650–1658. [Google Scholar] [CrossRef]
- Kuno, G.; Chang, G.J.; Tsuchiya, K.R.; Karabatsos, N.; Cropp, C.B. Phylogeny of the genus flavivirus. J. Virol. 1998, 72, 73–83. [Google Scholar] [CrossRef]
- Steinhauer, D.A.; Domingo, E.; Holland, J.J. Lack of evidence for proofreading mechanisms associated with an RNA virus polymerase. Gene 1992, 122, 281–288. [Google Scholar] [CrossRef]
- Drake, J.W. Rates of spontaneous mutation among RNA viruses. Proc. Natl. Acad. Sci. U. S. A. 1993, 90, 4171–4175. [Google Scholar] [CrossRef]
- Holmes, E.C. Patterns of intra- and interhost nonsynonymous variation reveal strong purifying selection in dengue virus. J. Virol. 2003, 77, 11296–11298. [Google Scholar] [CrossRef]
- Drummond, A.J.; Ho, S.Y.; Phillips, M.J.; Rambaut, A. Relaxed phylogenetics and dating with confidence. PLoS Biol. 2006, 4, e88. [Google Scholar] [CrossRef]
- Sall, A.A.; Faye, O.; Diallo, M.; Firth, C.; Kitchen, A.; Holmes, E.C. Yellow fever virus exhibits slower evolutionary dynamics than dengue virus. J. Virol. 2010, 84, 765–772. [Google Scholar] [CrossRef] [PubMed]
- Rosen, L. The emperor’s new clothes revisited, or reflections on the pathogenesis of dengue hemorrhagic fever. Am. J. Trop. Med. Hyg. 1977, 26, 337–343. [Google Scholar] [CrossRef] [PubMed]
- Lorono-Pino, M.A.; Cropp, C.B.; Farfan, J.A.; Vorndam, A.V.; Rodriguez-Angulo, E.M.; Rosado-Paredes, E.P.; Flores-Flores, L.F.; Beaty, B.J.; Gubler, D.J. Common occurrence of concurrent infections by multiple dengue virus serotypes. Am. J. Trop. Med. Hyg. 1999, 61, 725–730. [Google Scholar] [CrossRef]
- Holmes, E.C.; Burch, S.S. The causes and consequences of genetic variation in dengue virus. Trends Microbiol. 2000, 8, 74–77. [Google Scholar] [CrossRef] [PubMed]
- Scott, T.W.; Naksathit, A.; Day, J.F.; Kittayapong, P.; Edman, J.D. A fitness advantage for aedes aegypti and the viruses it transmits when females feed only on human blood. Am. J. Trop. Med. Hyg. 1997, 57, 235–239. [Google Scholar] [CrossRef] [PubMed]
- Craig, S.; Thu, H.M.; Lowry, K.; Wang, X.F.; Holmes, E.C.; Aaskov, J. Diverse dengue type 2 virus populations contain recombinant and both parental viruses in a single mosquito host. J. Virol. 2003, 77, 4463–4467. [Google Scholar] [CrossRef]
- Holmes, E.C.; Worobey, M.; Rambaut, A. Phylogenetic evidence for recombination in dengue virus. Mol. Biol. Evol. 1999, 16, 405–409. [Google Scholar] [CrossRef]
- Tolou, H.J.; Couissinier-Paris, P.; Durand, J.P.; Mercier, V.; de Pina, J.J.; de Micco, P.; Billoir, F.; Charrel, R.N.; de Lamballerie, X. Evidence for recombination in natural populations of dengue virus type 1 based on the analysis of complete genome sequences. J. Gen. Virol. 2001, 82, 1283–1290. [Google Scholar] [CrossRef]
- Uzcategui, N.Y.; Camacho, D.; Comach, G.; Cuello de Uzcategui, R.; Holmes, E.C.; Gould, E.A. Molecular epidemiology of dengue type 2 virus in Venezuela: Evidence for in situ virus evolution and recombination. J. Gen. Virol. 2001, 82, 2945–2953. [Google Scholar] [CrossRef]
- Worobey, M.; Rambaut, A.; Holmes, E.C. Widespread intra-serotype recombination in natural populations of dengue virus. Proc. Natl. Acad. Sci. U. S. A. 1999, 96, 7352–7357. [Google Scholar] [CrossRef]
- Twiddy, S.S.; Holmes, E.C. The extent of homologous recombination in members of the genus flavivirus. J. Gen. Virol. 2003, 84, 429–440. [Google Scholar] [CrossRef]
- Perez-Ramirez, G.; Diaz-Badillo, A.; Camacho-Nuez, M.; Cisneros, A.; Munoz Mde, L. Multiple recombinants in two dengue virus, serotype-2 isolates from patients from Oaxaca, Mexico. BMC Microbiol. 2009, 9, 260. [Google Scholar] [CrossRef] [PubMed]
- Aaskov, J.; Buzacott, K.; Field, E.; Lowry, K.; Berlioz-Arthaud, A.; Holmes, E.C. Multiple recombinant dengue type 1 viruses in an isolate from a dengue patient. J. Gen. Virol. 2007, 88, 3334–3340. [Google Scholar] [CrossRef]
- Chen, S.P.; Yu, M.; Jiang, T.; Deng, Y.Q.; Qin, C.F.; Han, J.F.; Qin, E.D. Identification of a recombinant dengue virus type 1 with 3 recombination regions in natural populations in Guangdong province, China. Arch. Virol. 2008, 153, 1175–1179. [Google Scholar] [CrossRef]
- Gubler, D.J. The global emergence/resurgence of arboviral diseases as public health problems. Arch. Med. Res. 2002, 33, 330–342. [Google Scholar] [CrossRef] [PubMed]
- Guzman, M.G.; Halstead, S.B.; Artsob, H.; Buchy, P.; Farrar, J.; Gubler, D.J.; Hunsperger, E.; Kroeger, A.; Margolis, H.S.; Martinez, E.; et al. Dengue: A continuing global threat. Nat. Microbiol. Rev. 2010, 8, S7–S16. [Google Scholar] [CrossRef]
- Carrillo-Valenzo, E.; Danis-Lozano, R.; Velasco-Hernandez, J.X.; Sanchez-Burgos, G.; Alpuche, C.; Lopez, I.; Rosales, C.; Baronti, C.; de Lamballerie, X.; Holmes, E.C.; Ramos-Castaneda, J. Evolution of dengue virus in mexico is characterized by frequent lineage replacement. Arch. Virol. 2010, 155, 1401–1412. [Google Scholar] [CrossRef] [PubMed]
- Scott, T.W.; Amerasinghe, P.H.; Morrison, A.C.; Lorenz, L.H.; Clark, G.G.; Strickman, D.; Kittayapong, P.; Edman, J.D. Longitudinal studies of aedes aegypti (diptera: Culicidae) in thailand and puerto rico: Blood feeding frequency. J. Med. Entomol. 2000, 37, 89–101. [Google Scholar] [CrossRef] [PubMed]
- Sanchez, I.J.; Ruiz, B.H. A single nucleotide change in the E protein gene of dengue virus 2 Mexican strain affects neurovirulence in mice. J. Gen. Virol. 1996, 77, 2541–2545. [Google Scholar] [CrossRef]
- Bennett, S.N.; Holmes, E.C.; Chirivella, M.; Rodriguez, D.M.; Beltran, M.; Vorndam, V.; Gubler, D.J.; McMillan, W.O. Molecular evolution of dengue 2 virus in Puerto Rico: Positive selection in the viral envelope accompanies clade reintroduction. J. Gen. Virol. 2006, 87, 885–893. [Google Scholar] [CrossRef]
- Mota, J.; Rico-Hesse, R. Humanized mice show clinical signs of dengue fever according to infecting virus genotype. J. Virol. 2009, 83, 8638–8645. [Google Scholar] [CrossRef] [PubMed]
- Lourenco, J.; Recker, M. Viral and epidemiological determinants of the invasion dynamics of novel dengue genotypes. PLoS Negl. Trop. Dis. 2010, 4, e894. [Google Scholar] [CrossRef] [PubMed]
- Guzman, M.G.; Kouri, G.; Halstead, S.B. Do escape mutants explain rapid increases in dengue case-fatality rates within epidemics? Lancet 2000, 355, 1902–1903. [Google Scholar] [CrossRef] [PubMed]
- Adams, B.; Holmes, E.C.; Zhang, C.; Mammen, M.P., Jr.; Nimmannitya, S.; Kalayanarooj, S.; Boots, M. Cross-protective immunity can account for the alternating epidemic pattern of dengue virus serotypes circulating in Bangkok. Proc. Natl. Acad. Sci. U. S. A. 2006, 103, 14234–14239. [Google Scholar] [CrossRef]
- Sittisombut, N.; Sistayanarain, A.; Cardosa, M.J.; Salminen, M.; Damrongdachakul, S.; Kalayanarooj, S.; Rojanasuphot, S.; Supawadee, J.; Maneekarn, N. Possible occurrence of a genetic bottleneck in dengue serotype 2 viruses between the 1980 and 1987 epidemic seasons in Bangkok, Thailand. Am. J. Trop. Med. Hyg. 1997, 57, 100–108. [Google Scholar] [CrossRef]
- Halstead, S.B.; Porterfield, J.S.; O’Rourke, E.J. Enhancement of dengue virus infection in monocytes by flavivirus antisera. Am. J. Trop. Med. Hyg. 1980, 29, 638–642. [Google Scholar] [CrossRef]
- Halstead, S.B. In vivo enhancement of dengue virus infection in rhesus monkeys by passively transferred antibody. J. Infect. Dis. 1979, 140, 527–533. [Google Scholar] [CrossRef]
- Halstead, S.B. Pathogenesis of dengue: Challenges to molecular biology. Science 1988, 239, 476–481. [Google Scholar] [CrossRef]
- Goncalvez, A.P.; Engle, R.E.; St Claire, M.; Purcell, R.H.; Lai, C.J. Monoclonal antibody-mediated enhancement of dengue virus infection in vitro and in vivo and strategies for prevention. Proc. Natl. Acad. Sci. U. S. A. 2007, 104, 9422–9427. [Google Scholar] [CrossRef]
- Cummings, D.A.; Schwartz, I.B.; Billings, L.; Shaw, L.B.; Burke, D.S. Dynamic effects of antibody-dependent enhancement on the fitness of viruses. Proc. Natl. Acad. Sci. U. S. A. 2005, 102, 15259–15264. [Google Scholar] [CrossRef]
- Holmes, E.C. The phylogeography of human viruses. Mol. Ecol. 2004, 13, 745–756. [Google Scholar] [CrossRef]
- Kawaguchi, I.; Sasaki, A.; Boots, M. Why are dengue virus serotypes so distantly related? Enhancement and limiting serotype similarity between dengue virus strains. Proc. Biol. Sci. 2003, 270, 2241–2247. [Google Scholar] [CrossRef]
- Adams, B.; Boots, M. Modelling the relationship between antibody-dependent enhancement and immunological distance with application to dengue. J. Theor. Biol. 2006, 242, 337–346. [Google Scholar] [CrossRef] [PubMed]
- Vasilakis, N.; Deardorff, E.R.; Kenney, J.L.; Rossi, S.L.; Hanley, K.A.; Weaver, S.C. Mosquitoes put the brake on arbovirus evolution: Experimental evolution reveals slower mutation accumulation in mosquito than vertebrate cells. PLoS Pathog. 2009, 5, e1000467. [Google Scholar] [CrossRef] [PubMed]
- Weaver, S.C.; Brault, A.C.; Kang, W.; Holland, J.J. Genetic and fitness changes accompanying adaptation of an arbovirus to vertebrate and invertebrate cells. J. Virol. 1999, 73, 4316–4326. [Google Scholar] [CrossRef] [PubMed]
- Greene, I.P.; Wang, E.; Deardorff, E.R.; Milleron, R.; Domingo, E.; Weaver, S.C. Effect of alternating passage on adaptation of sindbis virus to vertebrate and invertebrate cells. J. Virol. 2005, 79, 14253–14260. [Google Scholar] [CrossRef]
- Turner, P.E.; Elena, S.F. Cost of host radiation in an RNA virus. Genetics 2000, 156, 1465–1470. [Google Scholar] [CrossRef]
- Ciota, A.T.; Lovelace, A.O.; Ngo, K.A.; Le, A.N.; Maffei, J.G.; Franke, M.A.; Payne, A.F.; Jones, S.A.; Kauffman, E.B.; Kramer, L.D. Cell-specific adaptation of two flaviviruses following serial passage in mosquito cell culture. Virology 2007, 357, 165–174. [Google Scholar] [CrossRef]
- Ciota, A.T.; Lovelace, A.O.; Jia, Y.; Davis, L.J.; Young, D.S.; Kramer, L.D. Characterization of mosquito-adapted West Nile virus. J. Gen. Virol. 2008, 89, 1633–1642. [Google Scholar] [CrossRef]
- Coffey, L.L.; Vignuzzi, M. Host alteration in chikungunya virus increases fitness while restricting population diversity and adaptability to novel selective pressures. J. Virol. 2011, 85, 1025–1035. [Google Scholar] [CrossRef] [PubMed]
- Coffey, L.L.; Vasilakis, N.; Brault, A.C.; Powers, A.M.; Tripet, F.; Weaver, S.C. Arbovirus evolution in vivo is constrained by host alternation. Proc. Natl. Acad. Sci. U. S. A. 2008, 105, 6970–6975. [Google Scholar] [CrossRef] [PubMed]
- Lee, E.; Weir, R.C.; Dalgarno, L. Changes in the dengue virus major envelope protein on passaging and their localization on the three-dimensional structure of the protein. Virology 1997, 232, 281–290. [Google Scholar] [CrossRef]
- Novella, I.S.; Hershey, C.L.; Escarmis, C.; Domingo, E.; Holland, J.J. Lack of evolutionary stasis during alternating replication of an arbovirus in insect and mammalian cells. J. Mol. Biol. 1999, 287, 459–465. [Google Scholar] [CrossRef]
- Cooper, L.A.; Scott, T.W. Differential evolution of eastern equine encephalitis virus populations in response to host cell type. Genetics 2001, 157, 1403–1412. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Vargas, I.; Travanty, E.A.; Keene, K.M.; Franz, A.W.; Beaty, B.J.; Blair, C.D.; Olson, K.E. RNA interference, arthropod-borne viruses, and mosquitoes. Virus Res. 2004, 102, 65–74. [Google Scholar] [CrossRef]
- Chen, W.J.; Wu, H.R.; Chiou, S.S. E/NS1 modifications of dengue 2 virus after serial passages in mammalian and/or mosquito cells. Intervirology 2003, 46, 289–295. [Google Scholar] [CrossRef]
- de la Torre, J.C.; Holland, J.J. RNA virus quasispecies populations can suppress vastly superior mutant progeny. J. Virol. 1990, 64, 6278–6281. [Google Scholar] [CrossRef]
- Ciota, A.T.; Ngo, K.A.; Lovelace, A.O.; Payne, A.F.; Zhou, Y.; Shi, P.-Y.; Kramer, L.D. Role of the mutant spectrum in adaptation and replication of West Nile virus. J. Gen. Virol. 2007, 88, 865–874. [Google Scholar] [CrossRef] [PubMed]
- Ciota, A.T.; Lovelace, A.O.; Jones, S.A.; Payne, A.; Kramer, L.D. Adaptation of two flaviviruses results in differences in genetic heterogeneity and virus adaptability. J. Gen. Virol. 2007, 88, 2398–2406. [Google Scholar] [CrossRef]
- Jerzak, G.V.; Bernard, K.; Kramer, L.D.; Shi, P.Y.; Ebel, G.D. The West Nile virus mutant spectrum is host-dependant and a determinant of mortality in mice. Virology 2007, 360, 469–476. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2011 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Chen, R.; Vasilakis, N. Dengue — Quo tu et quo vadis? Viruses 2011, 3, 1562-1608. https://0-doi-org.brum.beds.ac.uk/10.3390/v3091562
Chen R, Vasilakis N. Dengue — Quo tu et quo vadis? Viruses. 2011; 3(9):1562-1608. https://0-doi-org.brum.beds.ac.uk/10.3390/v3091562
Chicago/Turabian StyleChen, Rubing, and Nikos Vasilakis. 2011. "Dengue — Quo tu et quo vadis?" Viruses 3, no. 9: 1562-1608. https://0-doi-org.brum.beds.ac.uk/10.3390/v3091562