2.1. Optimization of dot-ELISA method
The titer results of S10 polyclonal antibodies evaluated via indirect ELISA showed that both S10 multi-antibody titers reached 1:500 000, which was an excellent ratio. S10 antibody 1 (diluted ratio of 1:8000) had the highest OD value (2.187), whereas antibody 2 had the lowest (1.684). Then specificity of S10 polyclonal antibodies was measured by Western blot examination (
Figure 1), and we selected S10 polyclonal antibody 1 in our dot-ELISA assay. Preliminary experiments using viruliferous rice samples established the optimum dilutions of primary antibody needed for the dot-ELISA assay. Different dilutions of primary antibody in blocking buffer (1:1500 and 1:1000) and blocking time of primary incubation (60 min and 30 min) were tested (
Figure 2). Satisfactory results were obtained with a 1:1500 dilution of the primary following 30 min of blocking time. The NC membrane was used to test rice samples infected by SRBSDV (except in the seeding stage because of the influence of plant pigment) and exhibited the same diagnostic results as PVDF membrane.
Figure 1.
(A) Titers of the three SRBSDV S10 polyclonal antibodies. 1-3 represent antibodies 1, 2, 3 respectively. (B) Specificity analysis of the three SRBSDV S10 polyclonal antibodies 1, 2, 3 (from left to right).
Figure 1.
(A) Titers of the three SRBSDV S10 polyclonal antibodies. 1-3 represent antibodies 1, 2, 3 respectively. (B) Specificity analysis of the three SRBSDV S10 polyclonal antibodies 1, 2, 3 (from left to right).
Figure 2.
(A) 1:1000-fold antiserum dilution and 60 min second blocking time. (B) 1:1500-fold antiserum dilution and 60 min second blocking time. (C) 1:1500-fold antiserum dilution and 30 min second blocking time.
Figure 2.
(A) 1:1000-fold antiserum dilution and 60 min second blocking time. (B) 1:1500-fold antiserum dilution and 60 min second blocking time. (C) 1:1500-fold antiserum dilution and 30 min second blocking time.
2.2. Test with dot-ELISA method in lab
Our group tested hundreds of suspected SRBSDV rice samples from more than 60 districts in southwestern China from June to August 2011 (
Figure 3), and randomly chosen results were confirmed by the One Step RT-PCR assay. In the One Step RT-PCR assay with primers S10F/S10R, the unique band of SRBSDV occurred at 242 bp (
Figure 4).
Figure 3.
Distribution of SRBSDV infection in China (2011). Red indicates the samples infected with SRBSDV, the blue ones represent those uninfected.
Figure 3.
Distribution of SRBSDV infection in China (2011). Red indicates the samples infected with SRBSDV, the blue ones represent those uninfected.
Figure 4.
Positive controls were rice plants from Libo Guizhou province with confirmed SRBSDV infection. Negative controls were healthy rice plants grown in a greenhouse. (A) Dot-ELISA test (PVDF membrane) results of suspected rice plants from Shidian and Yuanjiang in Yunnan province. 1(+): Positive control. 2(+): Sample I from Shidian. 3(+): Sample II from Shidian. 4(+): Sample I from Yuanjiang. 5(+): Sample II from Yuanjiang. 6(-): Negative control. (B) Dot-ELISA test (PVDF membrane) results of suspected rice plants from Malipo Yunnan province. 1(+): Positive control. 2(+): Sample I. 3(+): Sample II. 4(+): Sample III. 5(-): Negative control. (C) One Step RT-PCR test results of suspected rice plants from Shidian and Yuanjiang. M: M, DL 500 bp DNA marker. 1: S10 gene amplification of rice sample I from Yuanjiang. 2: S10 gene amplification of rice sample IV from Shidian. 3: Reference gene amplification of rice sample I from Yuanjiang. 4: Reference gene amplification of rice sample IV from Shidian. 5: Multiple PCR of sample I from Yuanjiang. 6: Multiple PCR of sample IV from Shidian. 7: S10 gene amplification of positive sample. (D) One Step RT-PCR test results of suspected rice plants from Malipo. M: M, DL 500 bp DNA marker. 1: S10 gene amplification of rice sample I. 2: S10 gene amplification of rice sample II. 3: S10 gene amplification of rice sample III. 4: Positive control. 5: Reference gene amplification of rice sample I. 6: Reference gene amplification of rice sample II. 7: Reference gene amplification of rice sample III.
Figure 4.
Positive controls were rice plants from Libo Guizhou province with confirmed SRBSDV infection. Negative controls were healthy rice plants grown in a greenhouse. (A) Dot-ELISA test (PVDF membrane) results of suspected rice plants from Shidian and Yuanjiang in Yunnan province. 1(+): Positive control. 2(+): Sample I from Shidian. 3(+): Sample II from Shidian. 4(+): Sample I from Yuanjiang. 5(+): Sample II from Yuanjiang. 6(-): Negative control. (B) Dot-ELISA test (PVDF membrane) results of suspected rice plants from Malipo Yunnan province. 1(+): Positive control. 2(+): Sample I. 3(+): Sample II. 4(+): Sample III. 5(-): Negative control. (C) One Step RT-PCR test results of suspected rice plants from Shidian and Yuanjiang. M: M, DL 500 bp DNA marker. 1: S10 gene amplification of rice sample I from Yuanjiang. 2: S10 gene amplification of rice sample IV from Shidian. 3: Reference gene amplification of rice sample I from Yuanjiang. 4: Reference gene amplification of rice sample IV from Shidian. 5: Multiple PCR of sample I from Yuanjiang. 6: Multiple PCR of sample IV from Shidian. 7: S10 gene amplification of positive sample. (D) One Step RT-PCR test results of suspected rice plants from Malipo. M: M, DL 500 bp DNA marker. 1: S10 gene amplification of rice sample I. 2: S10 gene amplification of rice sample II. 3: S10 gene amplification of rice sample III. 4: Positive control. 5: Reference gene amplification of rice sample I. 6: Reference gene amplification of rice sample II. 7: Reference gene amplification of rice sample III.
![Viruses 04 00167 g004]()
Furthermore, the amplified SRBSDV genomic sequences from Shidian were analyzed (
Figure 5), and the homology of SRBSDV gene sequences between Shidian and Shaxian (retrieved from Genbank) in Fujian province was 99%. A purplish red dot illustrate that samples were infected with SRBSDV (
Figure 4). Based on comparisons, the dot-ELISA method was as accurate as the One Step RT-PCR assay for detecting infection in rice samples from Shidian, Yuanjiang and Malipo counties.
Figure 5.
Comparative genomic sequence of SRBSDV between Shidian and Shaxian.
Figure 5.
Comparative genomic sequence of SRBSDV between Shidian and Shaxian.
We also tested rice samples by dot-ELISA using PVDF membranes. Again the purple-red spots indicated that the tested rice strains from Lincang (one strain) and Chuxiong (
Figure 6) were infected with SRBSDV, while a sample from Menghai was not.
Figure 6.
Dot-ELISA test (PVDF membrane) results of suspected rice plants from Yunnan province. 1(-): Sample I from Menghai. 2(-): Sample II from Menghai. 3(-): Sample III from Menghai. 4(-): Sample IV from Menghai. 5(+): Sample from Shidian. 6(-): Sample I from Lincang. 7(+): Sample II from Lincang. 8(+): Sample from Chuxiong. 9(+): Positive control. 10(-): Negative control.
Figure 6.
Dot-ELISA test (PVDF membrane) results of suspected rice plants from Yunnan province. 1(-): Sample I from Menghai. 2(-): Sample II from Menghai. 3(-): Sample III from Menghai. 4(-): Sample IV from Menghai. 5(+): Sample from Shidian. 6(-): Sample I from Lincang. 7(+): Sample II from Lincang. 8(+): Sample from Chuxiong. 9(+): Positive control. 10(-): Negative control.
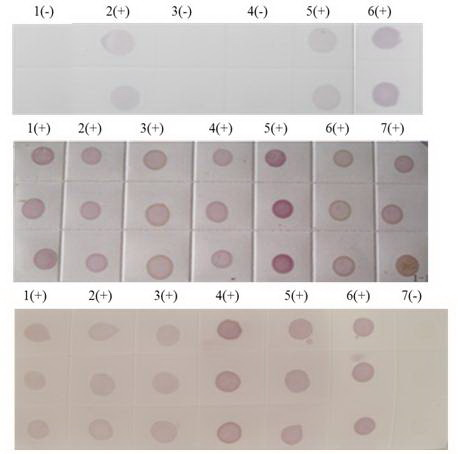
The sensitivities of PVDF and NC membrane were compared by parallel testing of samples from Tianzhu in Guizhou Province and Liangping in Chongqing City. The detection results of dot-ELISA were both positive (
Figure 7) but light purple spot indicated that the virus contents of the samples were lower than the positive control. Given that there was no difference in sensitivity between membranes, we selected the cheaper NC membrane for high throughput detection of SRBSDV infection.
Figure 7.
(A) Dot-ELISA test (PVDF membrane) results from Tianzhu Guizhou Province and Liangping Chongqing City. 1(+): Positive control. 2(+): Positive control. 3(+): Sample I from Tianzhu. 4(+): Sample II from Tianzhu. 5(+): Sample III from Tianzhu. 6(+): Sample from Liangping. 7(-): Negative control (stem). 8(-): Negative control (root). (B) Dot-ELISA test (NC membrane) results from Tianzhu and Liangping. 1(+): Positive control. 2(+): Positive control. 3(+): Sample I from Tianzhu. 4(+): Sample II from Tianzhu. 5(+): Sample III from Tianzhu. 6(+): Sample from Liangping. 7(-): Negative control (root). 8(-): Negative (stem).
Figure 7.
(A) Dot-ELISA test (PVDF membrane) results from Tianzhu Guizhou Province and Liangping Chongqing City. 1(+): Positive control. 2(+): Positive control. 3(+): Sample I from Tianzhu. 4(+): Sample II from Tianzhu. 5(+): Sample III from Tianzhu. 6(+): Sample from Liangping. 7(-): Negative control (stem). 8(-): Negative control (root). (B) Dot-ELISA test (NC membrane) results from Tianzhu and Liangping. 1(+): Positive control. 2(+): Positive control. 3(+): Sample I from Tianzhu. 4(+): Sample II from Tianzhu. 5(+): Sample III from Tianzhu. 6(+): Sample from Liangping. 7(-): Negative control (root). 8(-): Negative (stem).
The rice samples from 61 regions of Guizhou, Sichuan, and Yunnan province were tested in our laboratories, and 55 districts had at least one infected sample. Only selected results were discussed in this paper. The noticeable purple-positive spots of the samples from Luodian and Dushan in Guizhou province showed that these areas were experiencing an outbreak of SRBSDV disease (
Figure 8). In addition, samples from Kaiyuan, Jinghong, Shizong, Yongshan, Mile, Pingbian, Mengzi, Mengla, Shiping, Gejiu, Jianshui, Yiliang, Xinping, Fengqing, Shuifu, Zhenxiong, Yanjin, Maguan and Guangnan in Yunnan province were also positive for SRBSDV disease (
Figure 9). In contrast, there were no obvious spots on tests of samples from Huishui and Kaiyang in Guizhou province, Jiajiang in Sichuan province (
Figure 8), or Ludian and Kunming in Yunnan province (
Figure 9).
Figure 8.
Dot-ELISA test (NC membrane) results of suspected rice plants. (A) Luodian of Guizhou Province. 1(+): Positive control. 2(+): Sample from Dongjia in Luodian. 3(+): Sample from Bianyang in Luodian. 4(-): Negative control of root. 5(-): Negative control of stem. (B) Jiajiang of Sichuan province and Dushan of Guizhou Province. 1(+): Positive control. 2(-) - 7(-): Stem of sample from Jiajiang. 8(+): Sample from Dushan. 9(-): Negative control of root. 10(-): Negative control of stem. (C) Kaiyang and Huishui of Guizhou Province. 1(+): Positive control. 2(-): Root of sample from Kaiyang. 3(-): Stem sample from Kaiyang. 4(-): Stem of sample I from Huishui. 5(-): Root of Sample I from Huishui. 6(-): Root of Sample II rom Huishui. 7(-): Stem of sample II from Huishui. 8(-): Negative control of stem. 9 (-): Negative control of root.
Figure 8.
Dot-ELISA test (NC membrane) results of suspected rice plants. (A) Luodian of Guizhou Province. 1(+): Positive control. 2(+): Sample from Dongjia in Luodian. 3(+): Sample from Bianyang in Luodian. 4(-): Negative control of root. 5(-): Negative control of stem. (B) Jiajiang of Sichuan province and Dushan of Guizhou Province. 1(+): Positive control. 2(-) - 7(-): Stem of sample from Jiajiang. 8(+): Sample from Dushan. 9(-): Negative control of root. 10(-): Negative control of stem. (C) Kaiyang and Huishui of Guizhou Province. 1(+): Positive control. 2(-): Root of sample from Kaiyang. 3(-): Stem sample from Kaiyang. 4(-): Stem of sample I from Huishui. 5(-): Root of Sample I from Huishui. 6(-): Root of Sample II rom Huishui. 7(-): Stem of sample II from Huishui. 8(-): Negative control of stem. 9 (-): Negative control of root.
Figure 9.
Dot-ELISA test (NC membrane) results of suspected rice plants from Yunnanand Sichuan provinces. (A) 1(+) – 7(+): Kaiyuan, Jinghong, Shizong, Yongshan, Mile, Pingbian and Mengzi respectively. 8(-): Negative control of stem. 9(+): Positive control. (B) 1(+): Positive control. 9(+): Negative control of stem. 2(+) – 10(+): Mengla, Ludian, Gejiu, Yiliang, Fengqing, Shuifu, Zhenxiong, Yanjin. (C) 1(+): Positive control. 9(+): Negative control of stem. 2(+) – 10(+): Maguan, Jiangcheng, Eshan, Kunming, Qiubei, Yuxi, Fushun, Hejiang.
Figure 9.
Dot-ELISA test (NC membrane) results of suspected rice plants from Yunnanand Sichuan provinces. (A) 1(+) – 7(+): Kaiyuan, Jinghong, Shizong, Yongshan, Mile, Pingbian and Mengzi respectively. 8(-): Negative control of stem. 9(+): Positive control. (B) 1(+): Positive control. 9(+): Negative control of stem. 2(+) – 10(+): Mengla, Ludian, Gejiu, Yiliang, Fengqing, Shuifu, Zhenxiong, Yanjin. (C) 1(+): Positive control. 9(+): Negative control of stem. 2(+) – 10(+): Maguan, Jiangcheng, Eshan, Kunming, Qiubei, Yuxi, Fushun, Hejiang.
2.3. Diagnosis with dot-ELISA method in three county laboratories
During the same period, fast check-test technology for SRBSDV was applied in three county laboratories built by our group in Libo (Guizhou province), Shidian (Yunnan province) and Jianghua (Hunan province) as a means for early detection of SRBSDV outbreaks.
All samples picked from Libo, Shidian and Jianghua had the typical symptoms of SRBSDV disease, such as serious dwarf and wrinkled characteristic on leaves (
Figure 10). The results from the three county laboratories showed evident purple-positive spots, indicating the presence of SRBSDV disease in Libo, Shidian and Jianghua (
Figure 11). Furthermore, this fast check-test technology provided accurate and efficient data for local plant protection units to apply agriculture measures to control the spread of SRBSDV.
Figure 10.
Typical characterization of tillering stage rice plants infected with SRBSDV from three county laboratories.
Figure 10.
Typical characterization of tillering stage rice plants infected with SRBSDV from three county laboratories.
Figure 11.
Results in three county laboratories. (A) Libo of Guizhou province. 1(-): Negative control (stem tissue). 2(+) - 5(+): Suspected samples. 6(+): Positive control. (B) Shidian of Yunnan province. 1(+) - 6(+): Suspected samples collected from Shidian in Yunan province. 7(+): Positive control. (C) Jianghua of Hunan province. 1(+) - 5(+): Suspected samples. 6(+): Positive control. 7(-): Negative control (stem tissue).
Figure 11.
Results in three county laboratories. (A) Libo of Guizhou province. 1(-): Negative control (stem tissue). 2(+) - 5(+): Suspected samples. 6(+): Positive control. (B) Shidian of Yunnan province. 1(+) - 6(+): Suspected samples collected from Shidian in Yunan province. 7(+): Positive control. (C) Jianghua of Hunan province. 1(+) - 5(+): Suspected samples. 6(+): Positive control. 7(-): Negative control (stem tissue).
2.4. Discussion
At present, there is no effective and convenient method for SRBSDV detection in the field. Although PCR-based technologies reliably detect SRBSDV, these methods are time-consuming, laborious, and expensive, and impractical for local field detection. We developed an economical, quick and easy SRBSDV diagnosis kit. We obtained highly specific S10 polyclonal antibodies by the synthetic polypeptide technology, and measured them by Western blot experiments. Hence, a new rapid dot-ELISA method was improved with S10 polyclonal antibody 1. To further improve the reaction intensity from the background noise, different dilution ratios (1:1000 and 1:1500) in blocking buffer and incubation time were tested. Blocking the strips with 5% skim milk for 40 min at 37 °C produced the best results when SRBSDV antigen preparation was utilized. Therefore, for further testing, dot-ELISA was performed using SRBSDV antigen with 5% skim milk as the blocking agent to detect the SRBSDV antiserum. Of the different time periods tested for antiserum and conjugate (sheep anti-rabbit alkaline phosphatase), incubation of strips for 50 min in antiserum and 30 min in AP-conjugated secondary produced the best results. By means of these improvements, the assay for detection of SRBSDV infection could be finished within 3 hours.
In order to test the accuracy of dot-ELISA test, it was compared to the One Step RT-PCR method (
Figure 3). The tested rice samples were picked from Shidian county of Yunnan province, and these rice plants exhibited typical symptoms of infection, including stunting and dwarfing with stiff and wrinkled leaves. For the rice from Shidian, Yuan Jiang still in the tillering stage, the virus content was much lower as evidenced by the lighter purple spots; nonetheless, they were easily distinguished from the negative controls. This dot-ELISA method could accurately distinguish positive from negative samples as well as the One Step RT-PCR assay.
Both membranes (NC and PVDF) were equally suitable as revealed by direct comparison, but the NC membrane was less expensive, and so was used for most tests. The samples from Tianzhu in Guizhou Province and Liangping in Chongqing City were tested using both membranes in parallel experiments and both detected the presence of virus. In some cases, low levels of virus in seedling stage plants were likely to yield false-negatives, so it was necessary to use the higher specificity PVDF membrane. In some samples, the interference from chlorophyll, especially during the tillering stage of rice, interfered with the signal. In such cases, the blotted PVDF membranes could be washed in methanol with gently shaking for 15 to 30 seconds to remove most of the chlorophyll and other pigments to reduce the background interference.
This fast detection method was used in three county laboratories built in Libo (Guizhou province), Jianghua (Hunan province) and Shidian (Yunnan province), and these tests proved sufficiently accurate and sensitive for the early local detection of SRBSDV infections, allowing local officials to take appropriate responses to control spread. With the help of the early warning of SRBSDV infection, antiviral agent and insecticide were timely sprayed in local districts Libo, Jianghua and Shidian. The output results indicated decreased amounts of WBPHs and effectively controlled SRBSDV was ensured in these areas.
The causes and outbreak situation of SRBSDV infection were analyzed. Since 2010, serious SRBSDV outbreaks have occurred in Vietnam. The Chinese outbreaks may be due to WBPHs migrating from Vietnam, Myanmar, and Laos, all of which have had outbreaks. Guizhou, Sichuan, and especially Yunnan were hit by serious drought in 2011, which was particularly serious in central and eastern Yunan. Due to the drought and low winds, WBPH could not migrate, aggravating the SRBSDV outbreak in Yunnan in 2011. All bordering regions, such as Xuyong, Hejiang, Luodian, Congjiang, Guangnan, and Malipo, in addition to Chongqing city had outbreaks of SRBSDV dwarf disease (
Figure 2). The huge natural barrier formed by the surrounding mountains may prevent the WBPHs from migrating further, leading to local insect accumulation and SRBSDV outbreaks.
We report a new simple dot-ELISA method for the rapid detection of SRBSDV. All incubation steps are performed at 37 °C in a water bath shaker with gentle shaking, and the results can be seen by the naked eye, so precise instruments and trained researchers are not required. The test is applicable to diagnose infection in the field as well as in laboratories which are not well equipped. Compared to the One Step RT-PCR assay, the dot-ELISA method greatly simplifies the procedure, shortens the detection time, and saves on materials. It is simple enough for high throughput analysis of hundreds of samples per day. To further improve the sensitivity and specificity of this assay, investigations on developing higher avidity and specificity monoclonal antibodies are currently under way.