KSHV ORF57, a Protein of Many Faces
Abstract
:1. Introduction
2. The Primary Structure of ORF57 Protein

3. Expression and Localization of ORF57 Protein
4. Regulation of KSHV Gene Expression by ORF57
4.1. ORF57 Does Not Directly Export Viral RNAs

4.2. Stabilization of Viral Intronless Transcripts by ORF57
4.3. ORF57 Functions As a Viral Splicing Factor
4.4. ORF57 Promotes Protein Translation
4.5. Other Putative Functions of ORF57
5. Structural Determinants of ORF57 Function and Stability
5.1. ORF57 Secondary and Tertiary Structure


5.2. ORF57 Secondary Structure and Function
5.2.1. Roles of the ORF57 N-Terminal IDR in ORF57 Function
| ORF57 cofactor | Cofactor function | Interaction region in ORF57 (aa) | Interaction region in cofactor (aa/Domain) | Proved direct interaction | Proposed function of interaction | Conserved interaction in homologues | Citation | |
|---|---|---|---|---|---|---|---|---|
| Cellular cofactors | ||||||||
| Aly/REF | RNA-binding protein | 1–251 (181–215) | 103–163 (RRM) | + | RNA stability/Export (?) | + | [16,36] | |
| CBP80 | RNA-binding protein | ND | ND | − | RNA stability/Export (?)/Translation | − | [45,56] | |
| CKII α | Protein kinase | 181–455 | ND | + | Phopshorylation | + | [105] | |
| CKII α' | Protein kinase | 387–455 | ND | + | Phopshorylation | + | [105] | |
| CKII β | Protein kinase | 181–215 | 150-182 | + | Phopshorylation | + | [105] | |
| hnRNP K | RNA-binding protein | 17–181/329–387 | 240–337 (KI) interactive domain | + | ? | + | [105] | |
| hnRNP U | RNA-binding protein | 1–251 | ND | − | ? | − | [90] | |
| Nup62 | Nucleoporin | ND | ND | − | ? | + | [106] | |
| NXF1 | RNA export factor | ND | ND | − | Export (?) | + | [56] | |
| OTT3 | RNA-binding protein | 1–251 | 488–890 (SPOC) | + | RNA stability/Polyadenylation | + | [38] | |
| PABPC1 | RNA-binding protein | 1–251 | 1–370 (RRM1-4) | + | RNA stability | − | [56] | |
| PCBP1 | RNA-binding protein | 179–205 | 48–96 (KH-I) | + | Translation | − | [74] | |
| PYM | Scaffold protein | ND | ND | − | Translation | − | [75] | |
| RBM15 | RNA-binding protein | 1–251 | 530–977 (SPOC) | + | RNA stability/Polyadenylation | + | [38] | |
| SRSF1 | RNA-binding protein | ND | ND | − | RNA splicing | + | [20] | |
| SRSF3 | RNA-binding protein | 1–251 | 1–83 (RRM) | − | RNA splicing | + | [31] | |
| U snRNPs | Splicing factors | 1–251 | ND | − | RNA splicing | + | [20] | |
| U2AF35 | RNA-binding protein | 1–251 | ND | − | RNA splicing | + | [20] | |
| UAP56 | RNA-binding protein | ND | ND | − | Export (?) | + | [45] | |
| UBE2O | Ubiqitin-conjugating enzyme E2O | 1-251 | ND | - | ? | - | [90] | |
| UIF | RNA-binding protein | ND | ND | + | Export (?) | − | [50] | |
| Viral cofactors | ||||||||
| K-bZIP | Transcription factor | 96–215 | ND | + | ? | − | [85,86] | |
| ORF23 | ? | ND | ND | + | ? | − | [86] | |
| ORF50 | Transcription factor | ND | 1–518 (E3-LR) | + | Transactivation (?) | − | [83,84,86] | |
| ORF52 | ? | ND | ND | + | ? | − | [86] | |
| ORF57 | Posttranscriptional regulator | 251–455 | 251–455 | + | Self-interaction | + | [86,90,105] | |
| ORF61 | Ribonucleotide reductase | ND | ND | + | ? | + | [86] | |
| ORF63 | Tegument | ND | ND | + | ? | − | [86] | |
| ORF67.5 | ? | ND | ND | + | ? | + | [86] | |
| ORF68 | Glycoprotein | ND | ND | + | ? | + | [86] | |
5.2.2. Phosphorylation of the ORF57 N-Terminal Domain
5.2.3. Homodimerization of ORF57 via Its C-Terminus
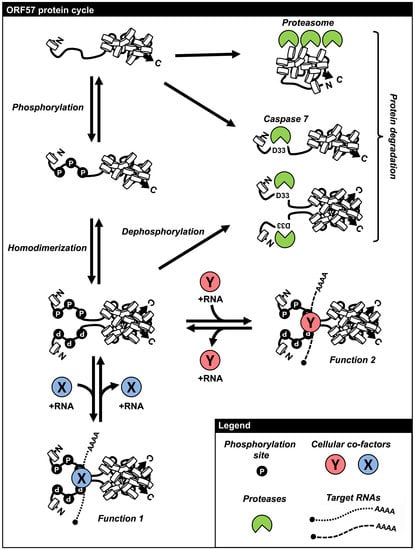
6. Regulation of ORF57 Stability by Phosphorylation and Homodimerization

7. Remarks and Perspectives
| ORF57 RNA target | Expression stage | Coding potential | Identified MRE element | ORF57 activity toward the target | Citation |
|---|---|---|---|---|---|
| Viral RNA targets | |||||
| vIL6 (K2) | E | Cytokine | MRE A/B | RNA stability/translation | [58] |
| K4# | E | Chemokine | ND | ? | [58] |
| K8 | E | Transcription factor (k-bZIP) | K8 intron 2 | Splicing | [20,31] |
| K8.1# | L | Glycoprotein | ND | ? | [8] |
| K9# | E | vIRF-1 | ND | ? | [58] |
| K12# | L | Kaposin | ND | ? | [58] |
| PAN | E | lncRNA | MRE I–III | RNA stability | [8,37,39,55] |
| mPC# | L | Minor capsid protein | ND | ? | [9] |
| ORF4 | E | Complement regulatory protein | ND | RNA stability (?) | [10] |
| ORF6 | E | Single-stranded DNA binding protein | ND | RNA stability (?) | [10] |
| ORF7# | L | Terminase subunit | ND | RNA stability (?) | [10] |
| ORF8# | L | Glycoprotein B | ND | RNA stability (?) | [10] |
| ORF9# | E | DNA polymerase | ND | ? | [9,10] |
| ORF17# | L | Viral protease | ND | ? | [58] |
| ORF47 | L | Glycoprotein | ND | RNA stability | [40,45] |
| ORF50 | IE | Transcription factor (Rta) | ND | Splicing | [20,58] |
| ORF52# | L | Tegument protein | ND | ? | [58] |
| ORF56 | E | Primase | ND | RNA stability | [22,40] |
| ORF59 | E | DNA polymerase processing factor | 5' MRE | RNA stability | [10,16,55,66] |
| ORF60# | E | Ribonucleotide reductase subunit | ND | RNA stability (?) | [10] |
| ORF61# | E | Ribonucleotide reductase subunit | ND | RNA stability (?) | [10] |
| ORF73# | L | LANA | ND | ? | [58] |
| Non-viral RNA targets | |||||
| hIL6 | N/A | Cytokine | miR-608 binding site | Translation | [58] |
| BPV-1 L1/L2 | N/A | Capsid proteins | SE4 | Splicing | [31] |
| HPV16 L1/L2 | N/A | Capsid proteins | ESE | Splicing | [31] |
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Nakamura, H.; Lu, M.; Gwack, Y.; Souvlis, J.; Zeichner, S.L.; Jung, J.U. Global changes in Kaposi’s sarcoma-associated virus gene expression patterns following expression of a tetracycline-inducible Rta transactivator. J. Virol. 2003, 77, 4205–4220. [Google Scholar] [CrossRef] [PubMed]
- Majerciak, V.; Ni, T.; Yang, W.; Meng, B.; Zhu, J.; Zheng, Z.M. A viral genome landscape of RNA polyadenylation from KSHV latent to lytic infection. PLoS Pathog. 2013, 9, e1003749. [Google Scholar] [CrossRef] [PubMed]
- Dresang, L.R.; Teuton, J.R.; Feng, H.; Jacobs, J.M.; Camp, D.G.; Purvine, S.O.; Gritsenko, M.A.; Li, Z.; Smith, R.D.; Sugden, B.; et al. Coupled transcriptome and proteome analysis of human lymphotropic tumor viruses: Insights on the detection and discovery of viral genes. BMC Genomics 2011, 12, 625. [Google Scholar] [CrossRef] [PubMed]
- Arias, C.; Weisburd, B.; Stern-Ginossar, N.; Mercier, A.; Madrid, A.S.; Bellare, P.; Holdorf, M.; Weissman, J.S.; Ganem, D. KSHV 2.0: A comprehensive annotation of the Kaposi’s sarcoma-associated herpesvirus genome using next-generation sequencing reveals novel genomic and functional features. PLoS Pathog. 2014, 10, e1003847. [Google Scholar] [CrossRef] [PubMed]
- Gradoville, L.; Gerlach, J.; Grogan, E.; Shedd, D.; Nikiforow, S.; Metroka, C.; Miller, G. Kaposi’s sarcoma-associated herpesvirus open reading frame 50/Rta protein activates the entire viral lytic cycle in the HH-B2 primary effusion lymphoma cell line. J. Virol. 2000, 74, 6207–6212. [Google Scholar] [CrossRef] [PubMed]
- Sun, R.; Lin, S.F.; Gradoville, L.; Yuan, Y.; Zhu, F.; Miller, G. A viral gene that activates lytic cycle expression of Kaposi’s sarcoma-associated herpesvirus. Proc. Natl. Acad. Sci. USA 1998, 95, 10866–10871. [Google Scholar] [CrossRef] [PubMed]
- Lukac, D.M.; Renne, R.; Kirshner, J.R.; Ganem, D. Reactivation of Kaposi’s sarcoma-associated herpesvirus infection from latency by expression of the ORF 50 transactivator, a homolog of the EBV R protein. Virology 1998, 252, 304–312. [Google Scholar] [CrossRef] [PubMed]
- Majerciak, V.; Pripuzova, N.; McCoy, J.P.; Gao, S.J.; Zheng, Z.M. Targeted disruption of Kaposi’s sarcoma-associated herpesvirus ORF57 in the viral genome is detrimental for the expression of ORF59, K8alpha, and K8.1 and the production of infectious virus. J. Virol. 2007, 81, 1062–1071. [Google Scholar] [CrossRef] [PubMed]
- Han, Z.; Swaminathan, S. Kaposi’s sarcoma-associated herpesvirus lytic gene ORF57 is essential for infectious virion production. J. Virol. 2006, 80, 5251–5260. [Google Scholar] [CrossRef] [PubMed]
- Verma, D.; Li, D.J.; Krueger, B.; Renne, R.; Swaminathan, S. Identification of the Physiological Gene Targets of the Essential Lytic Replicative Kaposi’s Sarcoma-Associated Herpesvirus ORF57 Protein. J. Virol. 2015, 89, 1688–1702. [Google Scholar] [CrossRef] [PubMed]
- Sandri-Goldin, R.M. The many roles of the regulatory protein ICP27 during herpes simplex virus infection. Front. Biosci. 2008, 13, 5241–5256. [Google Scholar] [CrossRef] [PubMed]
- Swaminathan, S. Post-transcriptional gene regulation by gamma herpesviruses. J. Cell. Biochem. 2005, 95, 698–711. [Google Scholar] [CrossRef] [PubMed]
- Boyne, J.R.; Colgan, K.J.; Whitehouse, A. Herpesvirus saimiri ORF57: A post-transcriptional regulatory protein. Front. Biosci. 2008, 13, 2928–2938. [Google Scholar] [CrossRef] [PubMed]
- Ote, I.; Lebrun, M.; Vandevenne, P.; Bontems, S.; Medina-Palazon, C.; Manet, E.; Piette, J.; Sadzot-Delvaux, C. Varicella-zoster virus IE4 protein interacts with SR proteins and exports mRNAs through the TAP/NXF1 pathway. PLoS One 2009, 4, e7882. [Google Scholar] [CrossRef] [PubMed]
- Majerciak, V.; Zheng, Z.M. Kaposi’s sarcoma-associated herpesvirus ORF57 in viral RNA processing. Front. Biosci. 2009, 14, 1516–1528. [Google Scholar] [CrossRef]
- Majerciak, V.; Yamanegi, K.; Nie, S.H.; Zheng, Z.M. Structural and functional analyses of Kaposi sarcoma-associated herpesvirus ORF57 nuclear localization signals in living cells. J. Biol. Chem. 2006, 281, 28365–28378. [Google Scholar] [CrossRef] [PubMed]
- Schenk, E.R.; Ridgeway, M.E.; Park, M.A.; Leng, F.; Fernandez-Lima, F. Isomerization kinetics of AT hook decapeptide solution structures. Anal. Chem. 2014, 86, 1210–1214. [Google Scholar] [CrossRef] [PubMed]
- Thandapani, P.; O’Connor, T.R.; Bailey, T.L.; Richard, S. Defining the RGG/RG motif. Mol. Cell 2013, 50, 613–623. [Google Scholar] [CrossRef] [PubMed]
- Moriuchi, M.; Moriuchi, H.; Debrus, S.; Piette, J.; Cohen, J.I. The acidic amino-terminal region of varicella-zoster virus open reading frame 4 protein is required for transactivation and can functionally replace the corresponding region of herpes simplex virus ICP27. Virology 1995, 208, 376–382. [Google Scholar] [CrossRef] [PubMed]
- Majerciak, V.; Yamanegi, K.; Allemand, E.; Kruhlak, M.; Krainer, A.R.; Zheng, Z.M. Kaposi’s sarcoma-associated herpesvirus ORF57 functions as a viral splicing factor and promotes expression of intron-containing viral lytic genes in spliceosome-mediated RNA splicing. J. Virol. 2008, 82, 2792–2801. [Google Scholar] [CrossRef] [PubMed]
- Li, D.J.; Verma, D.; Swaminathan, S. Binding of cellular export factor REF/Aly by Kaposi’s sarcoma-associated herpesvirus (KSHV) ORF57 protein is not required for efficient KSHV lytic replication. J. Virol. 2012, 86, 9866–9874. [Google Scholar] [CrossRef] [PubMed]
- Majerciak, V.; Yamanegi, K.; Zheng, Z.M. Gene structure and expression of Kaposi’s sarcoma-associated herpesvirus ORF56, ORF57, ORF58, and ORF59. J. Virol. 2006, 80, 11968–11981. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.; Chang, J.; Lynch, S.J.; Lukac, D.M.; Ganem, D. The lytic switch protein of KSHV activates gene expression via functional interaction with RBP-Jkappa (CSL), the target of the Notch signaling pathway. Genes Dev. 2002, 16, 1977–1989. [Google Scholar] [CrossRef] [PubMed]
- Izumiya, Y.; Izumiya, C.; Hsia, D.; Ellison, T.J.; Luciw, P.A.; Kung, H.J. NF-κB serves as a cellular sensor of Kaposi’s sarcoma-associated herpesvirus latency and negatively regulates K-Rta by antagonizing the RBP-Jkappa coactivator. J. Virol. 2009, 83, 4435–4446. [Google Scholar] [CrossRef] [PubMed]
- Wen, H.J.; Minhas, V.; Wood, C. Identification and characterization of a new Kaposi’s sarcoma-associated herpesvirus replication and transcription activator (RTA)-responsive element involved in RTA-mediated transactivation. J. Gen. Virol. 2009, 90, 944–953. [Google Scholar] [CrossRef] [PubMed]
- Vieira, J.; O’Hearn, P.M. Use of the red fluorescent protein as a marker of Kaposi’s sarcoma-associated herpesvirus lytic gene expression. Virology 2004, 325, 225–240. [Google Scholar] [CrossRef] [PubMed]
- Majerciak, V.; Deng, M.; Zheng, Z.M. Requirement of UAP56, URH49, RBM15, and OTT3 in the expression of Kaposi sarcoma-associated herpesvirus ORF57. Virology 2010, 407, 206–212. [Google Scholar] [CrossRef] [PubMed]
- Mears, W.E.; Lam, V.; Rice, S.A. Identification of nuclear and nucleolar localization signals in the herpes simplex virus regulatory protein ICP27. J. Virol. 1995, 69, 935–947. [Google Scholar] [PubMed]
- Hiriart, E.; Farjot, G.; Gruffat, H.; Nguyen, M.V.; Sergeant, A.; Manet, E. A novel nuclear export signal and a REF interaction domain both promote mRNA export by the Epstein-Barr virus EB2 protein. J. Biol. Chem. 2003, 278, 335–342. [Google Scholar] [CrossRef] [PubMed]
- Nekorchuk, M.; Han, Z.; Hsieh, T.T.; Swaminathan, S. Kaposi’s Sarcoma-Associated Herpesvirus ORF57 Protein Enhances mRNA Accumulation Independently of Effects on Nuclear RNA Export. J. Virol. 2007, 81, 9990–9998. [Google Scholar] [CrossRef] [PubMed]
- Majerciak, V.; Lu, M.; Li, X.; Zheng, Z.M. Attenuation of the suppressive activity of cellular splicing factor SRSF3 by Kaposi sarcoma-associated herpesvirus ORF57 protein is required for RNA splicing. RNA 2014, 20, 1747–1758. [Google Scholar] [CrossRef] [PubMed]
- Boyne, J.R.; Whitehouse, A. Nucleolar disruption impairs Kaposi’s sarcoma-associated herpesvirus ORF57-mediated nuclear export of intronless viral mRNAs. FEBS Lett. 2009, 583, 3549–3556. [Google Scholar] [CrossRef] [PubMed]
- Chen, I.H.; Sciabica, K.S.; Sandri-Goldin, R.M. ICP27 interacts with the RNA export factor Aly/REF to direct herpes simplex virus type 1 intronless mRNAs to the TAP export pathway. J. Virol. 2002, 76, 12877–12889. [Google Scholar] [CrossRef] [PubMed]
- Toth, Z.; Stamminger, T. The human cytomegalovirus regulatory protein UL69 and its effect on mRNA export. Front. Biosci. 2008, 13, 2939–2949. [Google Scholar] [CrossRef] [PubMed]
- Goodwin, D.J.; Hall, K.T.; Stevenson, A.J.; Markham, A.F.; Whitehouse, A. The open reading frame 57 gene product of herpesvirus saimiri shuttles between the nucleus and cytoplasm and is involved in viral RNA nuclear export. J. Virol. 1999, 73, 10519–10524. [Google Scholar] [PubMed]
- Malik, P.; Blackbourn, D.J.; Clements, J.B. The evolutionarily conserved Kaposi’s sarcoma-associated herpesvirus ORF57 protein interacts with REF protein and acts as an RNA export factor. J. Biol. Chem. 2004, 279, 33001–33011. [Google Scholar] [CrossRef] [PubMed]
- Massimelli, M.J.; Kang, J.G.; Majerciak, V.; Le, S.Y.; Liewehr, D.J.; Steinberg, S.M.; Zheng, Z.M. Stability of a Long Noncoding Viral RNA Depends on a 9-nt Core Element at the RNA 5' End to Interact with Viral ORF57 and Cellular PABPC1. Int. J. Biol. Sci. 2011, 7, 1145–1160. [Google Scholar] [CrossRef] [PubMed]
- Majerciak, V.; Uranishi, H.; Kruhlak, M.; Pilkington, G.R.; Massimelli, M.J.; Bear, J.; Pavlakis, G.N.; Felber, B.K.; Zheng, Z.M. Kaposi’s sarcoma-associated herpesvirus ORF57 interacts with cellular RNA export cofactors RBM15 and OTT3 to promote expression of viral ORF59. J. Virol. 2011, 85, 1528–1540. [Google Scholar] [CrossRef] [PubMed]
- Sahin, B.B.; Patel, D.; Conrad, N.K. Kaposi’s sarcoma-associated herpesvirus ORF57 protein binds and protects a nuclear noncoding RNA from cellular RNA decay pathways. PLoS Pathog. 2010, 6, e1000799. [Google Scholar] [CrossRef] [PubMed]
- Pilkington, G.R.; Majerciak, V.; Bear, J.; Uranishi, H.; Zheng, Z.M.; Felber, B.K. Kaposi’s Sarcoma-Associated Herpesvirus ORF57 Is Not a Bona Fide Export Factor. J. Virol. 2012, 86, 13089–13094. [Google Scholar] [CrossRef] [PubMed]
- Tunnicliffe, R.B.; Hautbergue, G.M.; Kalra, P.; Jackson, B.R.; Whitehouse, A.; Wilson, S.A.; Golovanov, A.P. Structural basis for the recognition of cellular mRNA export factor REF by herpes viral proteins HSV-1 ICP27 and HVS ORF57. PLoS Pathog. 2011, 7, e1001244. [Google Scholar] [CrossRef] [PubMed]
- Strasser, K.; Masuda, S.; Mason, P.; Pfannstiel, J.; Oppizzi, M.; Rodriguez-Navarro, S.; Rondon, A.G.; Aguilera, A.; Struhl, K.; Reed, R.; et al. TREX is a conserved complex coupling transcription with messenger RNA export. Nature 2002, 417, 304–308. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, J.P.; Rode, M.; Gatfield, D.; Blencowe, B.J.; Carmo-Fonseca, M.; Izaurralde, E. REF proteins mediate the export of spliced and unspliced mRNAs from the nucleus. Proc. Natl. Acad. Sci. USA 2001, 98, 1030–1035. [Google Scholar] [CrossRef] [PubMed]
- Nojima, T.; Hirose, T.; Kimura, H.; Hagiwara, M. The interaction between cap-binding complex and RNA export factor is required for intronless mRNA export. J. Biol. Chem. 2007, 282, 15645–15651. [Google Scholar] [CrossRef] [PubMed]
- Boyne, J.R.; Colgan, K.J.; Whitehouse, A. Recruitment of the complete hTREX complex is required for Kaposi’s sarcoma-associated herpesvirus intronless mRNA nuclear export and virus replication. PLoS Pathog. 2008, 4, e1000194. [Google Scholar] [CrossRef] [PubMed]
- Longman, D.; Johnstone, I.L.; Caceres, J.F. The Ref/Aly proteins are dispensable for mRNA export and development in Caenorhabditis elegans. RNA 2003, 9, 881–891. [Google Scholar] [CrossRef] [PubMed]
- Gatfield, D.; Izaurralde, E. REF1/Aly and the additional exon junction complex proteins are dispensable for nuclear mRNA export. J. Cell. Biol. 2002, 159, 579–588. [Google Scholar] [CrossRef] [PubMed]
- Johnson, L.A.; Li, L.; Sandri-Goldin, R.M. The cellular RNA export receptor TAP/NXF1 is required for ICP27-mediated export of herpes simplex virus 1 RNA, but the TREX complex adaptor protein Aly/REF appears to be dispensable. J. Virol. 2009, 83, 6335–6346. [Google Scholar] [CrossRef] [PubMed]
- Hautbergue, G.M.; Hung, M.L.; Walsh, M.J.; Snijders, A.P.; Chang, C.T.; Jones, R.; Ponting, C.P.; Dickman, M.J.; Wilson, S.A. UIF, a New mRNA export adaptor that works together with REF/ALY, requires FACT for recruitment to mRNA. Curr. Biol. 2009, 19, 1918–1924. [Google Scholar] [CrossRef] [PubMed]
- Jackson, B.R.; Boyne, J.R.; Noerenberg, M.; Taylor, A.; Hautbergue, G.M.; Walsh, M.J.; Wheat, R.; Blackbourn, D.J.; Wilson, S.A.; Whitehouse, A. An interaction between KSHV ORF57 and UIF provides mRNA-adaptor redundancy in herpesvirus intronless mRNA export. PLoS Pathog. 2011, 7, e1002138. [Google Scholar] [CrossRef] [PubMed]
- Stubbs, S.H.; Hunter, O.V.; Hoover, A.; Conrad, N.K. Viral factors reveal a role for REF/Aly in nuclear RNA stability. Mol. Cell. Biol. 2012, 32, 1260–1270. [Google Scholar] [CrossRef] [PubMed]
- Rossetto, C.C.; Pari, G. KSHV PAN RNA Associates with Demethylases UTX and JMJD3 to Activate Lytic Replication through a Physical Interaction with the Virus Genome. PLoS Pathog. 2012, 8, e1002680. [Google Scholar] [CrossRef] [PubMed]
- Rossetto, C.C.; Pari, G.S. KSHV noncoding PAN RNA interacts with virus and cellular-encoded proteins and suppresses expression of genes involved in immune modulation. J. Virol. 2011, 85, 13290–13297. [Google Scholar] [CrossRef] [PubMed]
- Campbell, M.; Kim, K.Y.; Chang, P.C.; Huerta, S.; Shevchenko, B.; Wang, D.H.; Izumiya, C.; Kung, H.J.; Izumiya, Y. A lytic viral long noncoding RNA modulates the function of a latent protein. J. Virol. 2014, 88, 1843–1848. [Google Scholar] [CrossRef] [PubMed]
- Kirshner, J.R.; Lukac, D.M.; Chang, J.; Ganem, D. Kaposi’s sarcoma-associated herpesvirus open reading frame 57 encodes a posttranscriptional regulator with multiple distinct activities. J. Virol. 2000, 74, 3586–3597. [Google Scholar] [CrossRef] [PubMed]
- Massimelli, M.J.; Majerciak, V.; Kruhlak, M.; Zheng, Z.M. Interplay between polyadenylate-binding protein 1 and Kaposi’s sarcoma-associated herpesvirus ORF57 in accumulation of polyadenylated nuclear RNA, a viral long noncoding RNA. J. Virol. 2013, 87, 243–256. [Google Scholar] [CrossRef] [PubMed]
- Sei, E.; Conrad, N.K. Delineation of a core RNA element required for Kaposi’s sarcoma-associated herpesvirus ORF57 binding and activity. Virology 2011, 419, 107–116. [Google Scholar] [CrossRef] [PubMed]
- Kang, J.G.; Pripuzova, N.; Majerciak, V.; Kruhlak, M.; Le, S.Y.; Zheng, Z.M. Kaposi’s Sarcoma-Associated Herpesvirus ORF57 Promotes Escape of Viral and Human Interleukin-6 from MicroRNA-Mediated Suppression. J. Virol. 2011, 85, 2620–2630. [Google Scholar] [CrossRef] [PubMed]
- Schwanhausser, B.; Busse, D.; Li, N.; Dittmar, G.; Schuchhardt, J.; Wolf, J.; Chen, W.; Selbach, M. Global quantification of mammalian gene expression control. Nature 2011, 473, 337–342. [Google Scholar] [CrossRef] [PubMed]
- Rabani, M.; Levin, J.Z.; Fan, L.; Adiconis, X.; Raychowdhury, R.; Garber, M.; Gnirke, A.; Nusbaum, C.; Hacohen, N.; Friedman, N.; et al. Metabolic labeling of RNA uncovers principles of RNA production and degradation dynamics in mammalian cells. Nat. Biotechnol. 2011, 29, 436–442. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tani, H.; Mizutani, R.; Salam, K.A.; Tano, K.; Ijiri, K.; Wakamatsu, A.; Isogai, T.; Suzuki, Y.; Akimitsu, N. Genome-wide determination of RNA stability reveals hundreds of short-lived noncoding transcripts in mammals. Genome Res. 2012, 22, 947–956. [Google Scholar] [CrossRef] [PubMed]
- Conrad, N.K.; Steitz, J.A. A Kaposi’s sarcoma virus RNA element that increases the nuclear abundance of intronless transcripts. EMBO J. 2005, 24, 1831–1841. [Google Scholar] [CrossRef] [PubMed]
- Conrad, N.K.; Shu, M.D.; Uyhazi, K.E.; Steitz, J.A. Mutational analysis of a viral RNA element that counteracts rapid RNA decay by interaction with the polyadenylate tail. Proc. Natl. Acad. Sci. USA 2007, 104, 10412–10417. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.J.; Glaunsinger, B.A. Aberrant herpesvirus-induced polyadenylation correlates with cellular messenger RNA destruction. PLoS Biol. 2009, 7, e1000107. [Google Scholar] [CrossRef] [PubMed]
- Borah, S.; Darricarrere, N.; Darnell, A.; Myoung, J.; Steitz, J.A. A Viral Nuclear Noncoding RNA Binds Re-localized Poly(A) Binding Protein and Is Required for Late KSHV Gene Expression. PLoS Pathog. 2011, 7, e1002300. [Google Scholar] [CrossRef] [PubMed]
- Massimelli, M.J.; Majerciak, V.; Kang, J.-G.; Liewehr, D.J.; Steinberg, S.M.; Zheng, Z.M. Multiple regions of Kaposi sarcoma-associated herpesvirus ORF59 RNA are required for its expression mediated by viral ORF57 and cellular RBM15. Viruses 2015, 7, 496–510. [Google Scholar] [CrossRef]
- Wu, X.; Brewer, G. The regulation of mRNA stability in mammalian cells: 2.0. Gene 2012, 500, 10–21. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Z.M. Split genes and their expression in Kaposi’s sarcoma-associated herpesvirus. Rev. Med. Virol. 2003, 13, 173–184. [Google Scholar] [CrossRef] [PubMed]
- Jia, R.; Li, C.; McCoy, J.P.; Deng, C.X.; Zheng, Z.M. SRp20 is a proto-oncogene critical for cell proliferation and tumor induction and maintenance. Int. J. Biol. Sci. 2010, 6, 806–826. [Google Scholar] [CrossRef] [PubMed]
- Verma, D.; Swaminathan, S. Epstein-Barr virus SM protein functions as an alternative splicing factor. J. Virol. 2008, 82, 7180–7188. [Google Scholar] [CrossRef] [PubMed]
- Verma, D.; Bais, S.; Gaillard, M.; Swaminathan, S. Epstein-Barr Virus SM protein utilizes cellular splicing factor SRp20 to mediate alternative splicing. J. Virol. 2010, 84, 11781–11789. [Google Scholar] [CrossRef] [PubMed]
- Sciabica, K.S.; Dai, Q.J.; Sandri-Goldin, R.M. ICP27 interacts with SRPK1 to mediate HSV splicing inhibition by altering SR protein phosphorylation. EMBO J. 2003, 22, 1608–1619. [Google Scholar] [CrossRef] [PubMed]
- Escudero-Paunetto, L.; Li, L.; Hernandez, F.P.; Sandri-Goldin, R.M. SR proteins SRp20 and 9G8 contribute to efficient export of herpes simplex virus 1 mRNAs. Virology 2010, 401, 155–164. [Google Scholar] [CrossRef] [PubMed]
- Nishimura, K.; Ueda, K.; Guwanan, E.; Sakakibara, S.; Do, E.; Osaki, E.; Yada, K.; Okuno, T.; Yamanishi, K. A posttranscriptional regulator of Kaposi’s sarcoma-associated herpesvirus interacts with RNA-binding protein PCBP1 and controls gene expression through the IRES. Virology 2004, 325, 364–378. [Google Scholar] [CrossRef] [PubMed]
- Boyne, J.R.; Jackson, B.R.; Taylor, A.; Macnab, S.A.; Whitehouse, A. Kaposi’s sarcoma-associated herpesvirus ORF57 protein interacts with PYM to enhance translation of viral intronless mRNAs. EMBO J. 2010, 29, 1851–1864. [Google Scholar] [CrossRef] [PubMed]
- Forler, D.; Kocher, T.; Rode, M.; Gentzel, M.; Izaurralde, E.; Wilm, M. An efficient protein complex purification method for functional proteomics in higher eukaryotes. Nat. Biotechnol. 2003, 21, 89–92. [Google Scholar] [CrossRef] [PubMed]
- Diem, M.D.; Chan, C.C.; Younis, I.; Dreyfuss, G. PYM binds the cytoplasmic exon-junction complex and ribosomes to enhance translation of spliced mRNAs. Nat. Struct. Mol. Biol. 2007, 14, 1173–1179. [Google Scholar] [CrossRef] [PubMed]
- Gehring, N.H.; Lamprinaki, S.; Kulozik, A.E.; Hentze, M.W. Disassembly of exon junction complexes by PYM. Cell 2009, 137, 536–548. [Google Scholar] [CrossRef] [PubMed]
- Kang, J.G.; Majerciak, V.; Uldrick, T.S.; Wang, X.; Kruhlak, M.; Yarchoan, R.; Zheng, Z.M. Kaposi’s sarcoma-associated herpesviral IL-6 and human IL-6 open reading frames contain miRNA binding sites and are subject to cellular miRNA regulation. J. Pathol. 2011, 225, 378–389. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, M.; Osborne, J.; Bestetti, G.; Chang, Y.; Moore, P.S. Viral IL-6-induced cell proliferation and immune evasion of interferon activity. Science 2002, 298, 1432–1435. [Google Scholar] [CrossRef] [PubMed]
- Jones, K.D.; Aoki, Y.; Chang, Y.; Moore, P.S.; Yarchoan, R.; Tosato, G. Involvement of interleukin-10 (IL-10) and viral IL-6 in the spontaneous growth of Kaposi’s sarcoma herpesvirus-associated infected primary effusion lymphoma cells. Blood 1999, 94, 2871–2879. [Google Scholar] [PubMed]
- Osborne, J.; Moore, P.S.; Chang, Y. KSHV-encoded viral IL-6 activates multiple human IL-6 signaling pathways. Hum. Immunol. 1999, 60, 921–927. [Google Scholar] [CrossRef] [PubMed]
- Malik, P.; Blackbourn, D.J.; Cheng, M.F.; Hayward, G.S.; Clements, J.B. Functional co-operation between the Kaposi’s sarcoma-associated herpesvirus ORF57 and ORF50 regulatory proteins. J. Gen. Virol. 2004, 85, 2155–2166. [Google Scholar] [CrossRef] [PubMed]
- Palmeri, D.; Spadavecchia, S.; Carroll, K.D.; Lukac, D.M. Promoter- and cell-specific transcriptional transactivation by the Kaposi’s sarcoma-associated herpesvirus ORF57/Mta protein. J. Virol. 2007, 81, 13299–13314. [Google Scholar] [CrossRef] [PubMed]
- Hunter, O.V.; Sei, E.; Richardson, R.B.; Conrad, N.K. Chromatin immunoprecipitation and microarray analysis suggest functional cooperation between Kaposi’s Sarcoma-associated herpesvirus ORF57 and K-bZIP. J. Virol. 2013, 87, 4005–4016. [Google Scholar] [CrossRef] [PubMed]
- Uetz, P.; Dong, Y.A.; Zeretzke, C.; Atzler, C.; Baiker, A.; Berger, B.; Rajagopala, S.V.; Roupelieva, M.; Rose, D.; Fossum, E.; et al. Herpesviral protein networks and their interaction with the human proteome. Science 2006, 311, 239–242. [Google Scholar] [CrossRef] [PubMed]
- Sandri-Goldin, R.M. The many roles of the highly interactive HSV protein ICP27, a key regulator of infection. Future Microbiol. 2011, 6, 1261–1277. [Google Scholar] [CrossRef] [PubMed]
- Dai-Ju, J.Q.; Li, L.; Johnson, L.A.; Sandri-Goldin, R.M. ICP27 interacts with the C-terminal domain of RNA polymerase II and facilitates its recruitment to herpes simplex virus 1 transcription sites, where it undergoes proteasomal degradation during infection. J. Virol. 2006, 80, 3567–3581. [Google Scholar] [CrossRef] [PubMed]
- Jackson, B.R.; Noerenberg, M.; Whitehouse, A. A novel mechanism inducing genome instability in Kaposi’s sarcoma-associated herpesvirus infected cells. PLoS Pathog. 2014, 10, e1004098. [Google Scholar] [CrossRef] [PubMed]
- Majerciak, V.; Pripuzova, N.; Chan, C.; Temkin, N.; Specht, S.I.; Zheng, Z.M. Stability of structured Kaposi sarcoma-associated herpesvirus ORF57 protein is regulated by protein phosphorylation and homodimerization. J. Virol. 2015. [Google Scholar] [CrossRef]
- Uversky, V.N.; Gillespie, J.R.; Fink, A.L. Why are “natively unfolded” proteins unstructured under physiologic conditions? Proteins 2000, 41, 415–427. [Google Scholar] [CrossRef] [PubMed]
- Lischka, P.; Thomas, M.; Toth, Z.; Mueller, R.; Stamminger, T. Multimerization of human cytomegalovirus regulatory protein UL69 via a domain that is conserved within its herpesvirus homologues. J. Gen. Virol. 2007, 88, 405–410. [Google Scholar] [CrossRef] [PubMed]
- Hernandez, F.P.; Sandri-Goldin, R.M. Herpes simplex virus 1 regulatory protein ICP27 undergoes a head-to-tail intramolecular interaction. J. Virol. 2010, 84, 4124–4135. [Google Scholar] [CrossRef] [PubMed]
- Taylor, A.; Jackson, B.R.; Noerenberg, M.; Hughes, D.J.; Boyne, J.R.; Verow, M.; Harris, M.; Whitehouse, A. Mutation of a C-terminal motif affects Kaposi’s sarcoma-associated herpesvirus ORF57 RNA binding, nuclear trafficking, and multimerization. J. Virol. 2011, 85, 7881–7891. [Google Scholar] [CrossRef] [PubMed]
- Jones, D.T. Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 1999, 292, 195–202. [Google Scholar] [CrossRef] [PubMed]
- Dosztanyi, Z.; Meszaros, B.; Simon, I. ANCHOR: Web server for predicting protein binding regions in disordered proteins. Bioinformatics 2009, 25, 2745–2746. [Google Scholar] [CrossRef] [PubMed]
- Iakoucheva, L.M.; Radivojac, P.; Brown, C.J.; O’Connor, T.R.; Sikes, J.G.; Obradovic, Z.; Dunker, A.K. The importance of intrinsic disorder for protein phosphorylation. Nucleic Acids Res. 2004, 32, 1037–1049. [Google Scholar] [CrossRef] [PubMed]
- Zhi, Y.; Sandri-Goldin, R.M. Analysis of the phosphorylation sites of herpes simplex virus type 1 regulatory protein ICP27. J. Virol. 1999, 73, 3246–3257. [Google Scholar] [PubMed]
- Medina-Palazon, C.; Gruffat, H.; Mure, F.; Filhol, O.; Vingtdeux-Didier, V.; Drobecq, H.; Cochet, C.; Sergeant, N.; Sergeant, A.; Manet, E. Protein kinase CK2 phosphorylation of EB2 regulates its function in the production of Epstein-Barr virus infectious viral particles. J. Virol. 2007, 81, 11850–11860. [Google Scholar] [CrossRef] [PubMed]
- Van der Lee, R.; Buljan, M.; Lang, B.; Weatheritt, R.J.; Daughdrill, G.W.; Dunker, A.K.; Fuxreiter, M.; Gough, J.; Gsponer, J.; Jones, D.T.; et al. Classification of intrinsically disordered regions and proteins. Chem. Rev. 2014, 114, 6589–6631. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dunker, A.K.; Silman, I.; Uversky, V.N.; Sussman, J.L. Function and structure of inherently disordered proteins. Curr. Opin. Struct. Biol. 2008, 18, 756–764. [Google Scholar] [CrossRef] [PubMed]
- Goh, G.K.; Dunker, A.K.; Uversky, V.N. Protein intrinsic disorder toolbox for comparative analysis of viral proteins. BMC Genomics 2008, 9 (Suppl. 2), S4. [Google Scholar] [CrossRef] [PubMed]
- Uversky, V.N.; Roman, A.; Oldfield, C.J.; Dunker, A.K. Protein intrinsic disorder and human papillomaviruses: Increased amount of disorder in E6 and E7 oncoproteins from high risk HPVs. J. Proteome Res. 2006, 5, 1829–1842. [Google Scholar] [CrossRef] [PubMed]
- Xue, B.; Blocquel, D.; Habchi, J.; Uversky, A.V.; Kurgan, L.; Uversky, V.N.; Longhi, S. Structural disorder in viral proteins. Chem. Rev. 2014, 114, 6880–6911. [Google Scholar] [CrossRef] [PubMed]
- Malik, P.; Clements, J.B. Protein kinase CK2 phosphorylation regulates the interaction of Kaposi’s sarcoma-associated herpesvirus regulatory protein ORF57 with its multifunctional partner hnRNP K. Nucleic Acids Res. 2004, 32, 5553–5569. [Google Scholar] [CrossRef] [PubMed]
- Malik, P.; Tabarraei, A.; Kehlenbach, R.H.; Korfali, N.; Iwasawa, R.; Graham, S.V.; Schirmer, E.C. Herpes simplex virus ICP27 protein directly interacts with the nuclear pore complex through Nup62, inhibiting host nucleocytoplasmic transport pathways. J. Biol. Chem. 2012, 287, 12277–12292. [Google Scholar] [CrossRef] [PubMed]
- Johnson, L.A.; Sandri-Goldin, R.M. Efficient nuclear export of herpes simplex virus 1 transcripts requires both RNA binding by ICP27 and ICP27 interaction with TAP/NXF1. J. Virol. 2009, 83, 1184–1192. [Google Scholar] [CrossRef] [PubMed]
- Toth, Z.; Lischka, P.; Stamminger, T. RNA-binding of the human cytomegalovirus transactivator protein UL69, mediated by arginine-rich motifs, is not required for nuclear export of unspliced RNA. Nucleic Acids Res. 2006, 34, 1237–1249. [Google Scholar] [CrossRef] [PubMed]
- Sergeant, A.; Gruffat, H.; Manet, E. The Epstein-Barr virus (EBV) protein EB is an mRNA export factor essential for virus production. Front. Biosci. 2008, 13, 3798–3813. [Google Scholar] [CrossRef] [PubMed]
- Tunnicliffe, R.B.; Hautbergue, G.M.; Wilson, S.A.; Kalra, P.; Golovanov, A.P. Competitive and cooperative interactions mediate RNA transfer from herpesvirus saimiri ORF57 to the mammalian export adaptor ALYREF. PLoS Pathog. 2014, 10, e1003907. [Google Scholar] [CrossRef] [PubMed]
- Calnan, B.J.; Biancalana, S.; Hudson, D.; Frankel, A.D. Analysis of arginine-rich peptides from the HIV Tat protein reveals unusual features of RNA-protein recognition. Genes Dev. 1991, 5, 201–210. [Google Scholar] [CrossRef] [PubMed]
- Lazinski, D.; Grzadzielska, E.; Das, A. Sequence-specific recognition of RNA hairpins by bacteriophage antiterminators requires a conserved arginine-rich motif. Cell 1989, 59, 207–218. [Google Scholar] [CrossRef] [PubMed]
- Bayer, T.S.; Booth, L.N.; Knudsen, S.M.; Ellington, A.D. Arginine-rich motifs present multiple interfaces for specific binding by RNA. RNA 2005, 11, 1848–1857. [Google Scholar] [CrossRef] [PubMed]
- Ruvolo, V.; Sun, L.; Howard, K.; Sung, S.; Delecluse, H.J.; Hammerschmidt, W.; Swaminathan, S. Functional analysis of Epstein-Barr virus SM protein: Identification of amino acids essential for structure, transactivation, splicing inhibition, and virion production. J. Virol. 2004, 78, 340–352. [Google Scholar] [CrossRef] [PubMed]
- Zhi, Y.; Sciabica, K.S.; Sandri-Goldin, R.M. Self-interaction of the herpes simplex virus type 1 regulatory protein ICP27. Virology 1999, 257, 341–351. [Google Scholar] [CrossRef] [PubMed]
- Majerciak, V.; Kruhlak, M.; Dagur, P.K.; McCoy, J.P., Jr.; Zheng, Z.M. Caspase-7 cleavage of Kaposi sarcoma-associated herpesvirus ORF57 confers a cellular function against viral lytic gene expression. J. Biol. Chem. 2010, 285, 11297–11307. [Google Scholar] [CrossRef] [PubMed]
- Desagher, S.; Osen-Sand, A.; Montessuit, S.; Magnenat, E.; Vilbois, F.; Hochmann, A.; Journot, L.; Antonsson, B.; Martinou, J.C. Phosphorylation of bid by casein kinases I and II regulates its cleavage by caspase 8. Mol. Cell 2001, 8, 601–611. [Google Scholar] [CrossRef] [PubMed]
- Tozser, J.; Bagossi, P.; Zahuczky, G.; Specht, S.I.; Majerova, E.; Copeland, T.D. Effect of caspase cleavage-site phosphorylation on proteolysis. Biochem. J. 2003, 372, 137–143. [Google Scholar] [CrossRef] [PubMed]
- Koffa, M.D.; Kean, J.; Zachos, G.; Rice, S.A.; Clements, J.B. CK2 protein kinase is stimulated and redistributed by functional herpes simplex virus ICP27 protein. J. Virol. 2003, 77, 4315–4325. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Majerciak, V.; Zheng, Z.-M. KSHV ORF57, a Protein of Many Faces. Viruses 2015, 7, 604-633. https://0-doi-org.brum.beds.ac.uk/10.3390/v7020604
Majerciak V, Zheng Z-M. KSHV ORF57, a Protein of Many Faces. Viruses. 2015; 7(2):604-633. https://0-doi-org.brum.beds.ac.uk/10.3390/v7020604
Chicago/Turabian StyleMajerciak, Vladimir, and Zhi-Ming Zheng. 2015. "KSHV ORF57, a Protein of Many Faces" Viruses 7, no. 2: 604-633. https://0-doi-org.brum.beds.ac.uk/10.3390/v7020604