Interactions between the HIV-1 Unspliced mRNA and Host mRNA Decay Machineries
Abstract
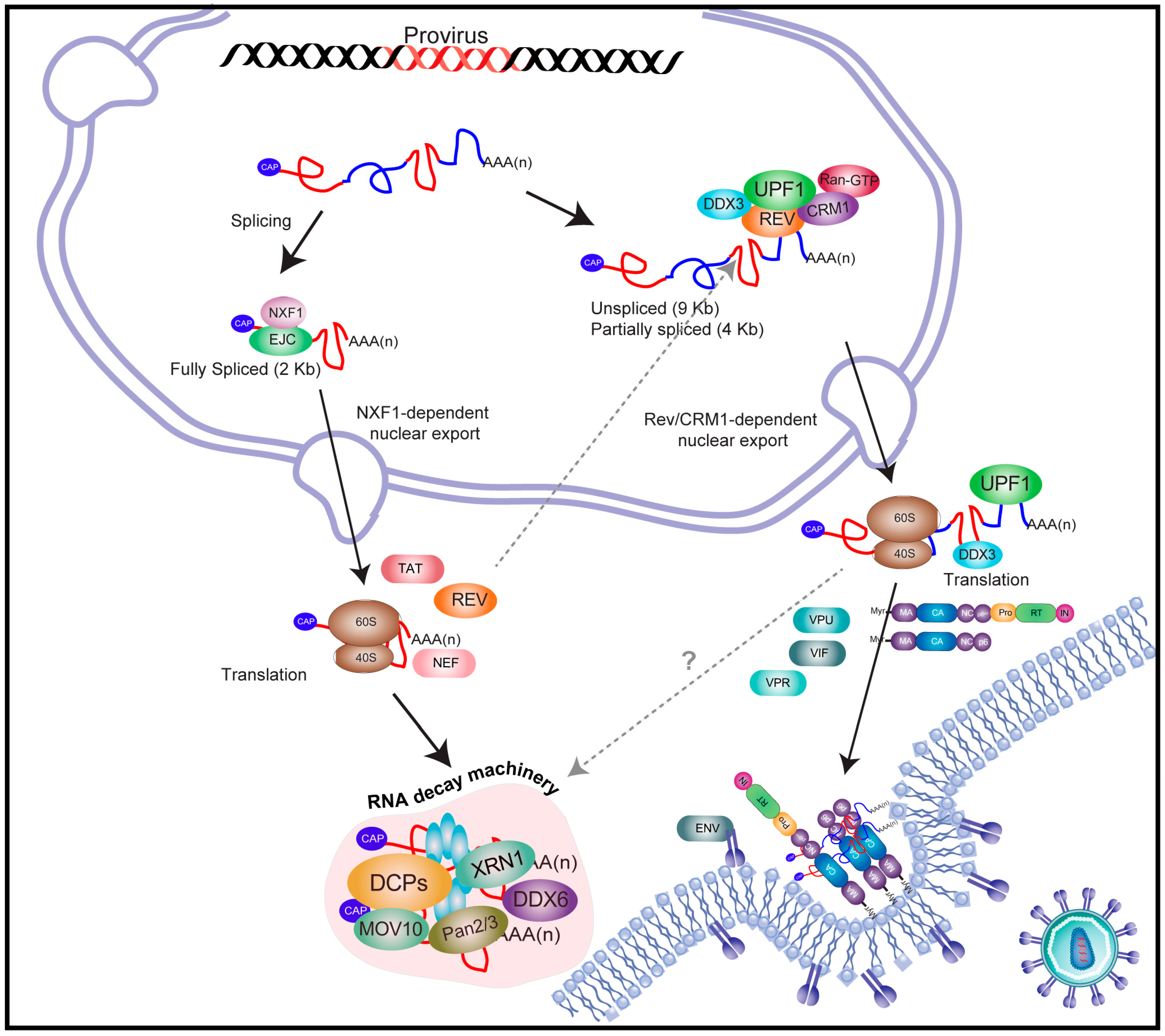
:1. Introduction
2. An Overview on Human Immunodeficiency Virus Type-1 (HIV-1) Gene Expression
3. The Full-Length and Partially Spliced mRNAs of HIV-1 Evade Nuclear Surveillance and Quality Control Cellular Mechanisms
4. REV Stabilizes RNA Instability Elements Present within the HIV-1 Unspliced mRNA
5. HIV-1 Recruits Factors Involved in Cytoplasmic mRNA Decay Pathways
5.1. Nonsense-Mediated Decay
5.2. Staufen-Mediated mRNA Decay
5.3. No-Go Decay
6. Relationship between HIV and the Cellular microRNA Machinery and Processing Bodies (PBs) Components
7. Interactions of HIV-1 and Components of RNA Granules Involved in mRNA Silencing
8. Control of HIV-1 mRNA Abundance by Methylation of Viral Transcripts
9. Conclusions and Future Perspectives
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Hogg, J.R. Viral evasion and manipulation of host RNA quality control pathways. J. Virol. 2016, 90, 7010–7018. [Google Scholar] [CrossRef] [PubMed]
- Davidson, L.; Kerr, A.; West, S. Co-transcriptional degradation of aberrant pre-mRNA by Xrn2. EMBO J. 2012, 31, 2566–2578. [Google Scholar] [CrossRef] [PubMed]
- Hilleren, P.; McCarthy, T.; Rosbash, M.; Parker, R.; Jensen, T.H. Quality control of mRNA 3′-end processing is linked to the nuclear exosome. Nature 2001, 413, 538–542. [Google Scholar] [CrossRef] [PubMed]
- Bousquet-Antonelli, C.; Presutti, C.; Tollervey, D. Identification of a regulated pathway for nuclear pre-mRNA turnover. Cell 2000, 102, 765–775. [Google Scholar] [CrossRef]
- Decker, C.J.; Parker, R. P-bodies and stress granules: Possible roles in the control of translation and mRNA degradation. Cold Spring Harb. Perspect. Biol. 2012, 4, a012286. [Google Scholar] [CrossRef] [PubMed]
- Doma, M.K.; Parker, R. Endonucleolytic cleavage of eukaryotic mRNAs with stalls in translation elongation. Nature 2006, 440, 561–564. [Google Scholar] [CrossRef] [PubMed]
- Harigaya, Y.; Parker, R. No-go decay: A quality control mechanism for RNA in translation. Wiley Interdiscip. Rev. RNA 2010, 1, 132–141. [Google Scholar] [CrossRef] [PubMed]
- Frischmeyer, P.A.; van Hoof, A.; O’Donnell, K.; Guerrerio, A.L.; Parker, R.; Dietz, H.C. An mRNA surveillance mechanism that eliminates transcripts lacking termination codons. Science 2002, 295, 2258–2261. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.Y.; Shyu, A.B. Selective degradation of early-response-gene mRNAs: Functional analyses of sequence features of the AU-rich elements. Mol. Cell. Biol. 1994, 14, 8471–8482. [Google Scholar] [CrossRef] [PubMed]
- Wilusz, C.J.; Wormington, M.; Peltz, S.W. The cap-to-tail guide to mRNA turnover. Nat. Rev. Mol. Cell Biol. 2001, 2, 237–246. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.Y.; Shyu, A.B. AU-rich elements: Characterization and importance in mRNA degradation. Trends Biochem. Sci. 1995, 20, 465–470. [Google Scholar] [CrossRef]
- Chen, C.Y.A.; Shyu, A.B. Emerging themes in regulation of global mRNA turnover in cis. Trends Biochem. Sci. 2016. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Zhao, B.S.; Roundtree, I.A.; Lu, Z.; Han, D.; Ma, H.; Weng, X.; Chen, K.; Shi, H.; He, C. N6-methyladenosine modulates messenger RNA translation efficiency. Cell 2015, 161, 1388–1399. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Lu, Z.; Gomez, A.; Hon, G.C.; Yue, Y.; Han, D.; Fu, Y.; Parisien, M.; Dai, Q.; Jia, G.; et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 2014, 505, 117–120. [Google Scholar] [CrossRef] [PubMed]
- Poblete-Duran, N.; Prades-Perez, Y.; Vera-Otarola, J.; Soto-Rifo, R.; Valiente-Echeverria, F. Who regulates whom? An overview of RNA granules and viral infections. Viruses 2016, 8, 180. [Google Scholar] [CrossRef] [PubMed]
- Cassan, E.; Arigon-Chifolleau, A.M.; Mesnard, J.M.; Gross, A.; Gascuel, O. Concomitant emergence of the antisense protein gene of HIV-1 and of the pandemic. Proc. Natl. Acad. Sci. USA 2016, 113, 11537–11542. [Google Scholar] [CrossRef] [PubMed]
- Frankel, A.D.; Young, J.A. HIV-1: Fifteen proteins and an RNA. Annu. Rev. Biochem. 1998, 67, 1–25. [Google Scholar] [CrossRef] [PubMed]
- Karn, J.; Stoltzfus, C.M. Transcriptional and posttranscriptional regulation of HIV-1 gene expression. Cold Spring Harb. Perspect. Med. 2012, 2, a006916. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.Y.; Byrn, R.; Groopman, J.; Baltimore, D. Temporal aspects of DNA and RNA synthesis during human immunodeficiency virus infection: Evidence for differential gene expression. J. Virol. 1989, 63, 3708–3713. [Google Scholar] [PubMed]
- Pomerantz, R.J.; Trono, D.; Feinberg, M.B.; Baltimore, D. Cells nonproductively infected with HIV-1 exhibit an aberrant pattern of viral RNA expression: A molecular model for latency. Cell 1990, 61, 1271–1276. [Google Scholar] [CrossRef]
- Purcell, D.F.; Martin, M.A. Alternative splicing of human immunodeficiency virus type 1 mRNA modulates viral protein expression, replication, and infectivity. J. Virol. 1993, 67, 6365–6378. [Google Scholar] [PubMed]
- Rojas-Araya, B.; Ohlmann, T.; Soto-Rifo, R. Translational control of the HIV unspliced genomic RNA. Viruses 2015, 7, 4326–4351. [Google Scholar] [CrossRef] [PubMed]
- Rajanala, K.; Nandicoori, V.K. Localization of nucleoporin Tpr to the nuclear pore complex is essential for Tpr mediated regulation of the export of unspliced RNA. PLoS ONE 2012, 7, e29921. [Google Scholar] [CrossRef] [PubMed]
- Coyle, J.H.; Bor, Y.C.; Rekosh, D.; Hammarskjold, M.L. The Tpr protein regulates export of mRNAs with retained introns that traffic through the Nxf1 pathway. RNA 2011, 17, 1344–1356. [Google Scholar] [CrossRef] [PubMed]
- Malim, M.H.; Hauber, J.; Le, S.Y.; Maizel, J.V.; Cullen, B.R. The HIV-1 Rev trans-activator acts through a structured target sequence to activate nuclear export of unspliced viral mRNA. Nature 1989, 338, 254–257. [Google Scholar] [CrossRef] [PubMed]
- Daly, T.J.; Cook, K.S.; Gray, G.S.; Maione, T.E.; Rusche, J.R. Specific binding of HIV-1 recombinant Rev protein to the Rev-Responsive Element in vitro. Nature 1989, 342, 816–819. [Google Scholar] [CrossRef] [PubMed]
- Fornerod, M.; Ohno, M.; Yoshida, M.; Mattaj, I.W. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell 1997, 90, 1051–1060. [Google Scholar] [CrossRef]
- Henderson, B.R.; Percipalle, P. Interactions between HIV Rev and nuclear import and export factors: The Rev nuclear localisation signal mediates specific binding to human importin-beta. J. Mol. Biol. 1997, 274, 693–707. [Google Scholar] [CrossRef] [PubMed]
- Arnold, M.; Nath, A.; Hauber, J.; Kehlenbach, R.H. Multiple Importins function as nuclear transport receptors for the Rev protein of human immunodeficiency virus type 1. J. Biol. Chem. 2006, 281, 20883–20890. [Google Scholar] [CrossRef] [PubMed]
- Fischer, U.; Huber, J.; Boelens, W.C.; Mattaj, I.W.; Luhrmann, R. The HIV-1 Rev activation domain is a nuclear export signal that accesses an export pathway used by specific cellular RNAs. Cell 1995, 82, 475–483. [Google Scholar] [CrossRef]
- Pollard, V.W.; Malim, M.H. The HIV-1 Rev protein. Annu. Rev. Microbiol. 1998, 52, 491–532. [Google Scholar] [CrossRef] [PubMed]
- Chang, D.D.; Sharp, P.A. Regulation by HIV Rev depends upon recognition of splice sites. Cell 1989, 59, 789–795. [Google Scholar] [CrossRef]
- Mann, D.A.; Mikaelian, I.; Zemmel, R.W.; Green, S.M.; Lowe, A.D.; Kimura, T.; Singh, M.; Butler, P.J.; Gait, M.J.; Karn, J. A molecular rheostat. Co-operative Rev binding to stem I of the Rev-response element modulates human immunodeficiency virus type-1 late gene expression. J. Mol. Biol. 1994, 241, 193–207. [Google Scholar] [CrossRef] [PubMed]
- Rausch, J.W.; Le Grice, S.F. HIV Rev assembly on the Rev response element (RRE): A structural perspective. Viruses 2015, 7, 3053–3075. [Google Scholar] [CrossRef] [PubMed]
- Cullen, B.R. Nuclear mRNA export: Insights from virology. Trends Biochem. Sci. 2003, 28, 419–424. [Google Scholar] [CrossRef]
- Suhasini, M.; Reddy, T.R. Cellular proteins and HIV-1 Rev function. Curr. HIV Res. 2009, 7, 91–100. [Google Scholar] [CrossRef] [PubMed]
- Kula, A.; Gharu, L.; Marcello, A. HIV-1 pre-mRNA commitment to Rev mediated export through PSF and Matrin 3. Virology 2013, 435, 329–340. [Google Scholar] [CrossRef] [PubMed]
- Jones, T.; Sheer, D.; Bevec, D.; Kappel, B.; Hauber, J.; Steinkasserer, A. The human HIV-1 Rev binding-protein hRIP/Rab (HRB) maps to chromosome 2q36. Genomics 1997, 40, 198–199. [Google Scholar] [CrossRef] [PubMed]
- Neville, M.; Stutz, F.; Lee, L.; Davis, L.I.; Rosbash, M. The importin-beta family member Crm1p bridges the interaction between Rev and the nuclear pore complex during nuclear export. Curr. Biol. 1997, 7, 767–775. [Google Scholar] [CrossRef]
- Yedavalli, V.S.; Neuveut, C.; Chi, Y.H.; Kleiman, L.; Jeang, K.T. Requirement of DDX3 DEAD box RNA helicase for HIV-1 Rev-RRE export function. Cell 2004, 119, 381–392. [Google Scholar] [CrossRef] [PubMed]
- Fang, J.; Kubota, S.; Yang, B.; Zhou, N.; Zhang, H.; Godbout, R.; Pomerantz, R.J. A DEAD box protein facilitates HIV-1 replication as a cellular co-factor of Rev. Virology 2004, 330, 471–480. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Tang, H.; Mullen, T.M.; Westberg, C.; Reddy, T.R.; Rose, D.W.; Wong-Staal, F. A role for RNA helicase A in post-transcriptional regulation of HIV type 1. Proc. Natl. Acad. Sci. USA 1999, 96, 709–714. [Google Scholar] [CrossRef] [PubMed]
- Felber, B.K.; Hadzopoulou-Cladaras, M.; Cladaras, C.; Copeland, T.; Pavlakis, G.N. Rev protein of human immunodeficiency virus type 1 affects the stability and transport of the viral mRNA. Proc. Natl. Acad. Sci. USA 1989, 86, 1495–1499. [Google Scholar] [CrossRef] [PubMed]
- Hadzopoulou-Cladaras, M.; Felber, B.K.; Cladaras, C.; Athanassopoulos, A.; Tse, A.; Pavlakis, G.N. The rev (trs/art) protein of human immunodeficiency virus type 1 affects viral mRNA and protein expression via a cis-acting sequence in the env region. J. Virol. 1989, 63, 1265–1274. [Google Scholar] [PubMed]
- Maldarelli, F.; Martin, M.A.; Strebel, K. Identification of posttranscriptionally active inhibitory sequences in human immunodeficiency virus type 1 RNA: Novel level of gene regulation. J. Virol. 1991, 65, 5732–5743. [Google Scholar] [PubMed]
- Schwartz, S.; Felber, B.K.; Pavlakis, G.N. Distinct RNA sequences in the gag region of human immunodeficiency virus type 1 decrease RNA stability and inhibit expression in the absence of Rev protein. J. Virol. 1992, 66, 150–159. [Google Scholar] [PubMed]
- Afonina, E.; Neumann, M.; Pavlakis, G.N. Preferential binding of poly(A)-binding protein 1 to an inhibitory RNA element in the human immunodeficiency virus type 1 gag mRNA. J. Biol. Chem. 1997, 272, 2307–2311. [Google Scholar] [PubMed]
- Nasioulas, G.; Zolotukhin, A.S.; Tabernero, C.; Solomin, L.; Cunningham, C.P.; Pavlakis, G.N.; Felber, B.K. Elements distinct from human immunodeficiency virus type 1 splice sites are responsible for the Rev dependence of env mRNA. J. Virol. 1994, 68, 2986–2993. [Google Scholar] [PubMed]
- Cochrane, A.W.; Jones, K.S.; Beidas, S.; Dillon, P.J.; Skalka, A.M.; Rosen, C.A. Identification and characterization of intragenic sequences which repress human immunodeficiency virus structural gene expression. J. Virol. 1991, 65, 5305–5313. [Google Scholar] [PubMed]
- Schwartz, S.; Campbell, M.; Nasioulas, G.; Harrison, J.; Felber, B.K.; Pavlakis, G.N. Mutational inactivation of an inhibitory sequence in human immunodeficiency virus type 1 results in Rev-independent gag expression. J. Virol. 1992, 66, 7176–7182. [Google Scholar] [PubMed]
- Zolotukhin, A.S.; Michalowski, D.; Bear, J.; Smulevitch, S.V.; Traish, A.M.; Peng, R.; Patton, J.; Shatsky, I.N.; Felber, B.K. PSF acts through the human immunodeficiency virus type 1 mRNA instability elements to regulate virus expression. Mol. Cell. Biol. 2003, 23, 6618–6630. [Google Scholar] [CrossRef] [PubMed]
- Schneider, R.; Campbell, M.; Nasioulas, G.; Felber, B.K.; Pavlakis, G.N. Inactivation of the human immunodeficiency virus type 1 inhibitory elements allows Rev-independent expression of Gag and Gag/protease and particle formation. J. Virol. 1997, 71, 4892–4903. [Google Scholar] [PubMed]
- Hentze, M.W. Determinants and regulation of cytoplasmic mRNA stability in eukaryotic cells. Biochim. Biophys. Acta 1991, 1090, 281–292. [Google Scholar] [CrossRef]
- Shyu, A.B.; Greenberg, M.E.; Belasco, J.G. The c-fos transcript is targeted for rapid decay by two distinct mRNA degradation pathways. Genes Dev. 1989, 3, 60–72. [Google Scholar] [CrossRef] [PubMed]
- Solomin, L.; Felber, B.K.; Pavlakis, G.N. Different sites of interaction for Rev, Tev, and Rex proteins within the Rev-Responsive Element of human immunodeficiency virus type 1. J. Virol. 1990, 64, 6010–6017. [Google Scholar] [PubMed]
- Shaw, G.; Kamen, R. A conserved AU sequence from the 3′ untranslated region of GM-CSF mRNA mediates selective mRNA degradation. Cell 1986, 46, 659–667. [Google Scholar] [CrossRef]
- Black, A.C.; Luo, J.; Watanabe, C.; Chun, S.; Bakker, A.; Fraser, J.K.; Morgan, J.P.; Rosenblatt, J.D. Polypyrimidine tract-binding protein and heterogeneous nuclear ribonucleoprotein A1 bind to human T-cell leukemia virus type 2 RNA regulatory elements. J. Virol. 1995, 69, 6852–6858. [Google Scholar] [PubMed]
- Black, A.C.; Luo, J.; Chun, S.; Bakker, A.; Fraser, J.K.; Rosenblatt, J.D. Specific binding of polypyrimidine tract binding protein and hnRNP a1 to HIV-1 CRS elements. Virus Genes 1996, 12, 275–285. [Google Scholar] [CrossRef] [PubMed]
- Valiente-Echeverria, F.; Vallejos, M.; Monette, A.; Pino, K.; Letelier, A.; Huidobro-Toro, J.P.; Mouland, A.J.; Lopez-Lastra, M. A cis-acting element present within the gag open reading frame negatively impacts on the activity of the HIV-1 IRES. PLoS ONE 2013, 8, e56962. [Google Scholar] [CrossRef] [PubMed]
- Maquat, L.E.; Tarn, W.Y.; Isken, O. The pioneer round of translation: Features and functions. Cell 2010, 142, 368–374. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Wilkinson, M.F. Regulation of nonsense-mediated mRNA decay. Wiley Interdiscip. Rev. RNA 2012, 3, 807–828. [Google Scholar] [CrossRef] [PubMed]
- Hoshino, S. Mechanism of the initiation of mRNA decay: Role of eRF3 family G proteins. Wiley Interdiscip. Rev. RNA 2012, 3, 743–757. [Google Scholar] [CrossRef] [PubMed]
- Schoenberg, D.R.; Maquat, L.E. Regulation of cytoplasmic mRNA decay. Nat. Rev. Genet. 2012, 13, 246–259. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, S.; Jacobson, A. RNA decay modulates gene expression and controls its fidelity. Wiley Interdiscip. Rev. RNA 2010, 1, 351–361. [Google Scholar] [CrossRef] [PubMed]
- Fabian, M.R.; Sonenberg, N. The mechanics of miRNA-mediated gene silencing: A look under the hood of miRISC. Nat. Struct. Mol. Biol. 2012, 19, 586–593. [Google Scholar] [CrossRef] [PubMed]
- Hwang, J.; Maquat, L.E. Nonsense-mediated mRNA decay (NMD) in animal embryogenesis: To die or not to die, that is the question. Curr. Opin. Genet. Dev. 2011, 21, 422–430. [Google Scholar] [CrossRef] [PubMed]
- Peltz, S.W.; Brown, A.H.; Jacobson, A. mRNA destabilization triggered by premature translational termination depends on at least three cis-acting sequence elements and one trans-acting factor. Genes Dev. 1993, 7, 1737–1754. [Google Scholar] [CrossRef] [PubMed]
- Schweingruber, C.; Rufener, S.C.; Zund, D.; Yamashita, A.; Muhlemann, O. Nonsense-mediated mRNA decay—Mechanisms of substrate mRNA recognition and degradation in mammalian cells. Biochim. Biophys. Acta 2013, 1829, 612–623. [Google Scholar] [CrossRef] [PubMed]
- Lykke-Andersen, J.; Shu, M.D.; Steitz, J.A. Human UPF proteins target an mRNA for nonsense-mediated decay when bound downstream of a termination codon. Cell 2000, 103, 1121–1131. [Google Scholar] [CrossRef]
- Serin, G.; Gersappe, A.; Black, J.D.; Aronoff, R.; Maquat, L.E. Identification and characterization of human orthologues to saccharomyces cerevisiae UPF2 protein and UPF3 protein (Caenorhabditis elegans SMG-4). Mol. Cell. Biol. 2001, 21, 209–223. [Google Scholar] [CrossRef] [PubMed]
- He, F.; Jacobson, A. Nonsense-mediated mRNA decay: Degradation of defective transcripts is only part of the story. Annu. Rev. Genet. 2015, 49, 339–366. [Google Scholar] [CrossRef] [PubMed]
- Karousis, E.D.; Nasif, S.; Muhlemann, O. Nonsense-mediated mRNA decay: Novel mechanistic insights and biological impact. Wiley Interdiscip. Rev. RNA 2016, 7, 661–682. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Balistreri, G.; Horvath, P.; Schweingruber, C.; Zund, D.; McInerney, G.; Merits, A.; Muhlemann, O.; Azzalin, C.; Helenius, A. The host nonsense-mediated mRNA decay pathway restricts mammalian RNA virus replication. Cell Host Microb. 2014, 16, 403–411. [Google Scholar] [CrossRef] [PubMed]
- Ajamian, L.; Abrahamyan, L.; Milev, M.; Ivanov, P.V.; Kulozik, A.E.; Gehring, N.H.; Mouland, A.J. Unexpected roles for UPF1 in HIV-1 RNA metabolism and translation. RNA 2008, 14, 914–927. [Google Scholar] [CrossRef] [PubMed]
- Hogg, J.R.; Goff, S.P. Upf1 senses 3′ UTR length to potentiate mRNA decay. Cell 2010, 143, 379–389. [Google Scholar] [CrossRef] [PubMed]
- Ajamian, L.; Abel, K.; Rao, S.; Vyboh, K.; Garcia-de-Gracia, F.; Soto-Rifo, R.; Kulozik, A.E.; Gehring, N.H.; Mouland, A.J. HIV-1 recruits UPF1 but excludes UPF2 to promote nucleocytoplasmic export of the genomic RNA. Biomolecules 2015, 5, 2808–2839. [Google Scholar] [CrossRef] [PubMed]
- Serquina, A.K.; Das, S.R.; Popova, E.; Ojelabi, O.A.; Roy, C.K.; Gottlinger, H.G. UPF1 is crucial for the infectivity of human immunodeficiency virus type 1 progeny virions. J. Virol. 2013, 87, 8853–8861. [Google Scholar] [CrossRef] [PubMed]
- Mocquet, V.; Durand, S.; Jalinot, P. How Retroviruses escape the Nonsense-Mediated mRNA Decay. AIDS Res. Hum. Retrovir. 2015, 31, 948–958. [Google Scholar] [CrossRef] [PubMed]
- Cullen, B.R. Role and mechanism of action of the APOBECc3 family of antiretroviral resistance factors. J. Virol. 2006, 80, 1067–1076. [Google Scholar] [CrossRef] [PubMed]
- Köhrmann, M.; Luo, M.; Kaether, C.; DesGroseillers, L.; Dotti, C.G.; Kiebler, M.A. Microtubule-dependent recruitment of Staufen-green fluorescent protein into large RNA-containing granules and subsequent dendritic transport in living hippocampal neurons. Mol. Biol. Cell 1999, 10, 2945–2953. [Google Scholar] [CrossRef] [PubMed]
- Kiebler, M.A.; Hemraj, I.; Verkade, P.; Köhrmann, M.; Fortes, P.; Marión, R.M.; Ortı́n, J.; Dotti, C.G. The mammalian staufen protein localizes to the somatodendritic domain of cultured hippocampal neurons: Implications for its involvement in mRNA transport. J. Neurosci. 1999, 19, 288–297. [Google Scholar] [PubMed]
- Ricci, E.P.; Kucukural, A.; Cenik, C.; Mercier, B.C.; Singh, G.; Heyer, E.E.; Ashar-Patel, A.; Peng, L.; Moore, M.J. Staufen1 senses overall transcript secondary structure to regulate translation. Nat. Struct. Mol. Biol. 2014, 21, 26–35. [Google Scholar] [CrossRef] [PubMed]
- Sugimoto, Y.; Vigilante, A.; Darbo, E.; Zirra, A.; Militti, C.; D’Ambrogio, A.; Luscombe, N.M.; Ule, J. hiCLIP reveals the in vivo atlas of mRNA secondary structures recognized by Staufen 1. Nature 2015, 519, 491–494. [Google Scholar] [CrossRef] [PubMed]
- Park, E.; Maquat, L.E. Staufen-mediated mRNA decay. Wiley Interdiscip. Rev. RNA 2013, 4, 423–435. [Google Scholar] [CrossRef] [PubMed]
- Dugre-Brisson, S.; Elvira, G.; Boulay, K.; Chatel-Chaix, L.; Mouland, A.J.; DesGroseillers, L. Interaction of Staufen1 with the 5′ end of mRNA facilitates translation of these RNAs. Nucleic Acids Res. 2005, 33, 4797–4812. [Google Scholar] [CrossRef] [PubMed]
- Chatel-Chaix, L.; Clement, J.F.; Martel, C.; Beriault, V.; Gatignol, A.; DesGroseillers, L.; Mouland, A.J. Identification of Staufen in the human immunodeficiency virus type 1 Gag ribonucleoprotein complex and a role in generating infectious viral particles. Mol. Cell. Biol. 2004, 24, 2637–2648. [Google Scholar] [CrossRef] [PubMed]
- Chatel-Chaix, L.; Abrahamyan, L.; Frechina, C.; Mouland, A.J.; DesGroseillers, L. The host protein Staufen1 participates in human immunodeficiency virus type 1 assembly in live cells by influencing pr55Gag multimerization. J. Virol. 2007, 81, 6216–6230. [Google Scholar] [CrossRef] [PubMed]
- Chatel-Chaix, L.; Boulay, K.; Mouland, A.J.; Desgroseillers, L. The host protein Staufen1 interacts with the pr55Gag zinc fingers and regulates HIV-1 assembly via its N-terminus. Retrovirology 2008, 5, 41. [Google Scholar] [CrossRef] [PubMed]
- Abrahamyan, L.G.; Chatel-Chaix, L.; Ajamian, L.; Milev, M.P.; Monette, A.; Clement, J.F.; Song, R.; Lehmann, M.; DesGroseillers, L.; Laughrea, M.; et al. Novel Staufen1 ribonucleoproteins prevent formation of stress granules but favour encapsidation of HIV-1 genomic RNA. J. Cell Sci. 2010, 123, 369–383. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, A.; Benjamin, R.; Balakrishnan, K.; Ghosh, P.; Banerjee, S. Human protein Staufen-2 promotes HIV-1 proliferation by positively regulating RNA export activity of viral protein Rev. Retrovirology 2014, 11, 18. [Google Scholar] [CrossRef] [PubMed]
- Mu, X.; Fu, Y.; Zhu, Y.; Wang, X.; Xuan, Y.; Shang, H.; Goff, S.P.; Gao, G. HIV-1 exploits the host factor RuvB-like 2 to balance viral protein expression. Cell Host Microb. 2015, 18, 233–242. [Google Scholar] [CrossRef] [PubMed]
- Voinnet, O. RNA silencing: Small RNAs as ubiquitous regulators of gene expression. Curr. Opin. Plant Biol. 2002, 5, 444–451. [Google Scholar] [CrossRef]
- Saumet, A.; Lecellier, C.H. Anti-viral RNA silencing: Do we look like plants? Retrovirology 2006, 3, 3. [Google Scholar] [CrossRef] [PubMed]
- Kim, V.N. MicroRNA biogenesis: Coordinated cropping and dicing. Nat. Rev. Mol. Cell Biol. 2005, 6, 376–385. [Google Scholar] [CrossRef] [PubMed]
- Nathans, R.; Chu, C.Y.; Serquina, A.K.; Lu, C.C.; Cao, H.; Rana, T.M. Cellular microRNA and P bodies modulate host-HIV-1 interactions. Mol. Cell 2009, 34, 696–709. [Google Scholar] [CrossRef] [PubMed]
- Pillai, R.S.; Bhattacharyya, S.N.; Filipowicz, W. Repression of protein synthesis by miRNAs: How many mechanisms? Trends Cell Biol. 2007, 17, 118–126. [Google Scholar] [CrossRef] [PubMed]
- Haase, A.D.; Jaskiewicz, L.; Zhang, H.; Laine, S.; Sack, R.; Gatignol, A.; Filipowicz, W. TRBP, a regulator of cellular PKR and HIV-1 virus expression, interacts with Dicer and functions in RNA silencing. EMBO Rep. 2005, 6, 961–967. [Google Scholar] [CrossRef] [PubMed]
- Triboulet, R.; Mari, B.; Lin, Y.L.; Chable-Bessia, C.; Bennasser, Y.; Lebrigand, K.; Cardinaud, B.; Maurin, T.; Barbry, P.; Baillat, V.; et al. Suppression of microRNA-silencing pathway by HIV-1 during virus replication. Science 2007, 315, 1579–1582. [Google Scholar] [CrossRef] [PubMed]
- Hayashita, Y.; Osada, H.; Tatematsu, Y.; Yamada, H.; Yanagisawa, K.; Tomida, S.; Yatabe, Y.; Kawahara, K.; Sekido, Y.; Takahashi, T. A polycistronic microRNA cluster, miR-17–92, is overexpressed in human lung cancers and enhances cell proliferation. Cancer Res. 2005, 65, 9628–9632. [Google Scholar] [CrossRef] [PubMed]
- Tagawa, H.; Seto, M. A microRNA cluster as a target of genomic amplification in malignant lymphoma. Leukemia 2005, 19, 2013–2016. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Thomson, J.M.; Hemann, M.T.; Hernando-Monge, E.; Mu, D.; Goodson, S.; Powers, S.; Cordon-Cardo, C.; Lowe, S.W.; Hannon, G.J.; et al. A microRNA polycistron as a potential human oncogene. Nature 2005, 435, 828–833. [Google Scholar] [CrossRef] [PubMed]
- Gregersen, L.H.; Schueler, M.; Munschauer, M.; Mastrobuoni, G.; Chen, W.; Kempa, S.; Dieterich, C.; Landthaler, M. MOV10 is a 5′ to 3′ RNA helicase contributing to UPF1 mRNA target degradation by translocation along 3′ UTRs. Mol. Cell 2014, 54, 573–585. [Google Scholar] [CrossRef] [PubMed]
- Abudu, A.; Wang, X.; Dang, Y.; Zhou, T.; Xiang, S.H.; Zheng, Y.H. Identification of molecular determinants from moloney leukemia virus 10 homolog (MOV10) protein for virion packaging and anti-HIV-1 activity. J. Biol. Chem. 2012, 287, 1220–1228. [Google Scholar] [CrossRef] [PubMed]
- Gallois-Montbrun, S.; Kramer, B.; Swanson, C.M.; Byers, H.; Lynham, S.; Ward, M.; Malim, M.H. Antiviral protein APOBEC3G localizes to ribonucleoprotein complexes found in P bodies and stress granules. J. Virol. 2007, 81, 2165–2178. [Google Scholar] [CrossRef] [PubMed]
- Burdick, R.; Smith, J.L.; Chaipan, C.; Friew, Y.; Chen, J.; Venkatachari, N.J.; Delviks-Frankenberry, K.A.; Hu, W.S.; Pathak, V.K. P body-associated protein MOV10 inhibits HIV-1 replication at multiple stages. J. Virol. 2010, 84, 10241–10253. [Google Scholar] [CrossRef] [PubMed]
- Huang, F.; Zhang, J.; Zhang, Y.; Geng, G.; Liang, J.; Li, Y.; Chen, J.; Liu, C.; Zhang, H. RNA helicase MOV10 functions as a co-factor of HIV-1 Rev to facilitate Rev/RRE-dependent nuclear export of viral mRNAs. Virology 2015, 486, 15–26. [Google Scholar] [CrossRef] [PubMed]
- Ingelfinger, D.; Arndt-Jovin, D.J.; Luhrmann, R.; Achsel, T. The human LSm1–7 proteins colocalize with the mRNA-degrading enzymes DCP1/2 and Xrnl in distinct cytoplasmic foci. RNA 2002, 8, 1489–1501. [Google Scholar] [PubMed]
- Yu, J.H.; Yang, W.H.; Gulick, T.; Bloch, K.D.; Bloch, D.B. Ge-1 is a central component of the mammalian cytoplasmic mRNA processing body. RNA 2005, 11, 1795–1802. [Google Scholar] [CrossRef] [PubMed]
- Sheth, U.; Parker, R. Targeting of aberrant mRNAs to cytoplasmic processing bodies. Cell 2006, 125, 1095–1109. [Google Scholar] [CrossRef] [PubMed]
- Durand, S.; Cougot, N.; Mahuteau-Betzer, F.; Nguyen, C.H.; Grierson, D.S.; Bertrand, E.; Tazi, J.; Lejeune, F. Inhibition of nonsense-mediated mRNA decay (NMD) by a new chemical molecule reveals the dynamic of NMD factors in P-bodies. J. Cell Biol. 2007, 178, 1145–1160. [Google Scholar] [CrossRef] [PubMed]
- Wilczynska, A.; Aigueperse, C.; Kress, M.; Dautry, F.; Weil, D. The translational regulator CPEB1 provides a link between DCP1 bodies and stress granules. J. Cell Sci. 2005, 118, 981–992. [Google Scholar] [CrossRef] [PubMed]
- Andrei, M.A.; Ingelfinger, D.; Heintzmann, R.; Achsel, T.; Rivera-Pomar, R.; Luhrmann, R. A role for eIF4e and eIF4e-transporter in targeting mRNPs to mammalian processing bodies. RNA 2005, 11, 717–727. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Rivas, F.V.; Wohlschlegel, J.; Yates, J.R., 3rd; Parker, R.; Hannon, G.J. A role for the P-body component GW182 in microRNA function. Nat. Cell Biol. 2005, 7, 1261–1266. [Google Scholar] [CrossRef] [PubMed]
- Pillai, R.S.; Bhattacharyya, S.N.; Artus, C.G.; Zoller, T.; Cougot, N.; Basyuk, E.; Bertrand, E.; Filipowicz, W. Inhibition of translational initiation by Let-7 microRNA in human cells. Science 2005, 309, 1573–1576. [Google Scholar] [CrossRef] [PubMed]
- Sen, G.L.; Blau, H.M. Argonaute 2/RISC resides in sites of mammalian mRNA decay known as cytoplasmic bodies. Nat. Cell Biol. 2005, 7, 633–636. [Google Scholar] [CrossRef] [PubMed]
- Eulalio, A.; Behm-Ansmant, I.; Izaurralde, E. P bodies: At the crossroads of post-transcriptional pathways. Nat. Rev. Mol. Cell Biol. 2007, 8, 9–22. [Google Scholar] [CrossRef] [PubMed]
- Bouttier, M.; Saumet, A.; Peter, M.; Courgnaud, V.; Schmidt, U.; Cazevieille, C.; Bertrand, E.; Lecellier, C.H. Retroviral GAG proteins recruit AGO2 on viral RNAs without affecting RNA accumulation and translation. Nucleic Acids Res. 2012, 40, 775–786. [Google Scholar] [CrossRef] [PubMed]
- Reed, J.C.; Molter, B.; Geary, C.D.; McNevin, J.; McElrath, J.; Giri, S.; Klein, K.C.; Lingappa, J.R. HIV-1 Gag co-opts a cellular complex containing DDX6, a helicase that facilitates capsid assembly. J. Cell Biol. 2012, 198, 439–456. [Google Scholar] [CrossRef] [PubMed]
- Zimmerman, C.; Klein, K.C.; Kiser, P.K.; Singh, A.R.; Firestein, B.L.; Riba, S.C.; Lingappa, J.R. Identification of a host protein essential for assembly of immature HIV-1 capsids. Nature 2002, 415, 88–92. [Google Scholar] [CrossRef] [PubMed]
- Lingappa, J.R.; Dooher, J.E.; Newman, M.A.; Kiser, P.K.; Klein, K.C. Basic residues in the nucleocapsid domain of Gag are required for interaction of HIV-1 Gag with ABCE1 (HP68), a cellular protein important for HIV-1 capsid assembly. J. Biol. Chem. 2006, 281, 3773–3784. [Google Scholar] [CrossRef] [PubMed]
- Kedersha, N.; Stoecklin, G.; Ayodele, M.; Yacono, P.; Lykke-Andersen, J.; Fritzler, M.J.; Scheuner, D.; Kaufman, R.J.; Golan, D.E.; Anderson, P. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J. Cell Biol. 2005, 169, 871–884. [Google Scholar] [CrossRef] [PubMed]
- Kedersha, N.L.; Gupta, M.; Li, W.; Miller, I.; Anderson, P. RNA-binding proteins TIA-1 and TIAR link the phosphorylation of eIF-2 alpha to the assembly of mammalian stress granules. J. Cell Biol. 1999, 147, 1431–1442. [Google Scholar] [CrossRef] [PubMed]
- Kedersha, N.; Cho, M.R.; Li, W.; Yacono, P.W.; Chen, S.; Gilks, N.; Golan, D.E.; Anderson, P. Dynamic shuttling of TIA-1 accompanies the recruitment of mRNA to mammalian stress granules. J. Cell Biol. 2000, 151, 1257–1268. [Google Scholar] [CrossRef] [PubMed]
- Valiente-Echeverria, F.; Melnychuk, L.; Vyboh, K.; Ajamian, L.; Gallouzi, I.E.; Bernard, N.; Mouland, A.J. eEF2 and Ras-GAP SH3 domain-binding protein (G3BP1) modulate stress granule assembly during HIV-1 infection. Nat. Commun. 2014, 5, 4819. [Google Scholar] [CrossRef] [PubMed]
- Cinti, A.; Le Sage, V.; Ghanem, M.; Mouland, A.J. HIV-1 Gag blocks selenite-induced stress granule assembly by altering the mRNA cap-binding complex. mBio 2016, 7, e00329. [Google Scholar] [CrossRef] [PubMed]
- Cobos Jimenez, V.; Martinez, F.O.; Booiman, T.; van Dort, K.A.; van de Klundert, M.A.; Gordon, S.; Geijtenbeek, T.B.; Kootstra, N.A. G3BP1 restricts HIV-1 replication in macrophages and T-cells by sequestering viral RNA. Virology 2015, 486, 94–104. [Google Scholar] [CrossRef] [PubMed]
- Cantara, W.A.; Crain, P.F.; Rozenski, J.; McCloskey, J.A.; Harris, K.A.; Zhang, X.; Vendeix, F.A.; Fabris, D.; Agris, P.F. The RNA modification database, RNAMDB: 2011 Update. Nucleic Acids Res. 2011, 39, D195–D201. [Google Scholar] [CrossRef] [PubMed]
- Du, H.; Zhao, Y.; He, J.; Zhang, Y.; Xi, H.; Liu, M.; Ma, J.; Wu, L. YTHDF2 destabilizes m6A-containing RNA through direct recruitment of the CCR4-NOT deadenylase complex. Nat. Commun. 2016, 7, 12626. [Google Scholar] [CrossRef] [PubMed]
- Beemon, K.; Keith, J. Localization of N6-methyladenosine in the rous sarcoma virus genome. J. Mol. Biol. 1977, 113, 165–179. [Google Scholar] [CrossRef]
- Canaani, D.; Kahana, C.; Lavi, S.; Groner, Y. Identification and mapping of N6-methyladenosine containing sequences in simian virus 40 RNA. Nucleic Acids Res. 1979, 6, 2879–2899. [Google Scholar] [CrossRef] [PubMed]
- Hashimoto, S.I.; Green, M. Multiple methylated cap sequences in adenovirus type 2 early mRNA. J. Virol. 1976, 20, 425–435. [Google Scholar] [PubMed]
- Krug, R.M.; Morgan, M.A.; Shatkin, A.J. Influenza viral mRNA contains internal N6-methyladenosine and 5′-terminal 7-methylguanosine in cap structures. J. Virol. 1976, 20, 45–53. [Google Scholar] [PubMed]
- Moss, B.; Gershowitz, A.; Stringer, J.R.; Holland, L.E.; Wagner, E.K. 5′-terminal and internal methylated nucleosides in herpes simplex virus type 1 mRNA. J. Virol. 1977, 23, 234–239. [Google Scholar] [PubMed]
- Kennedy, E.M.; Bogerd, H.P.; Kornepati, A.V.; Kang, D.; Ghoshal, D.; Marshall, J.B.; Poling, B.C.; Tsai, K.; Gokhale, N.S.; Horner, S.M.; et al. Post-transcriptional m6A editing of HIV-1 mRNAs enhances viral gene expression. Cell Host Microb. 2016, 19, 675–685. [Google Scholar] [CrossRef] [PubMed]
- Lichinchi, G.; Gao, S.; Saletore, Y.; Gonzalez, G.M.; Bansal, V.; Wang, Y.; Mason, C.E.; Rana, T.M. Dynamics of the human and viral m6A RNA methylomes during HIV-1 infection of t cells. Nat. Microbiol. 2016, 1, 16011. [Google Scholar] [CrossRef] [PubMed]
- Tirumuru, N.; Zhao, B.S.; Lu, W.; Lu, Z.; He, C.; Wu, L. N6-methyladenosine of HIV-1 RNA regulates viral infection and HIV-1 Gag protein expression. eLife 2016, 5, e15528. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Yue, Y.; Han, D.; Wang, X.; Fu, Y.; Zhang, L.; Jia, G.; Yu, M.; Lu, Z.; Deng, X.; et al. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat. Chem. Biol. 2014, 10, 93–95. [Google Scholar] [CrossRef] [PubMed]
- Meyer, K.D.; Saletore, Y.; Zumbo, P.; Elemento, O.; Mason, C.E.; Jaffrey, S.R. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell 2012, 149, 1635–1646. [Google Scholar] [CrossRef] [PubMed]
- Yue, Y.; Liu, J.; He, C. RNA N6-methyladenosine methylation in post-transcriptional gene expression regulation. Genes Dev. 2015, 29, 1343–1355. [Google Scholar] [CrossRef] [PubMed]
- Jia, G.; Fu, Y.; Zhao, X.; Dai, Q.; Zheng, G.; Yang, Y.; Yi, C.; Lindahl, T.; Pan, T.; Yang, Y.G.; et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 2011, 7, 885–887. [Google Scholar] [CrossRef] [PubMed]
- Zheng, G.; Dahl, J.A.; Niu, Y.; Fedorcsak, P.; Huang, C.M.; Li, C.J.; Vagbo, C.B.; Shi, Y.; Wang, W.L.; Song, S.H.; et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol. Cell 2013, 49, 18–29. [Google Scholar] [CrossRef] [PubMed]
- Xiao, W.; Adhikari, S.; Dahal, U.; Chen, Y.S.; Hao, Y.J.; Sun, B.F.; Sun, H.Y.; Li, A.; Ping, X.L.; Lai, W.Y.; et al. Nuclear m6A reader YTHDC1 regulates mRNA splicing. Mol. Cell 2016, 61, 507–519. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Zhao, D.; Wu, J.; Shi, Y. Structure of the YTH domain of human YTHDF2 in complex with an m6A mononucleotide reveals an aromatic cage for m6A recognition. Cell Res. 2014, 24, 1490–1492. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; He, C. Reading RNA methylation codes through methyl-specific binding proteins. RNA Biol. 2014, 11, 669–672. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Li, Y.; Toth, J.I.; Petroski, M.D.; Zhang, Z.; Zhao, J.C. N6-methyladenosine modification destabilizes developmental regulators in embryonic stem cells. Nat. Cell Biol. 2014, 16, 191–198. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.; Dai, Q.; Zheng, G.; He, C.; Parisien, M.; Pan, T. N6-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature 2015, 518, 560–564. [Google Scholar] [CrossRef] [PubMed]

© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Toro-Ascuy, D.; Rojas-Araya, B.; Valiente-Echeverría, F.; Soto-Rifo, R. Interactions between the HIV-1 Unspliced mRNA and Host mRNA Decay Machineries. Viruses 2016, 8, 320. https://0-doi-org.brum.beds.ac.uk/10.3390/v8110320
Toro-Ascuy D, Rojas-Araya B, Valiente-Echeverría F, Soto-Rifo R. Interactions between the HIV-1 Unspliced mRNA and Host mRNA Decay Machineries. Viruses. 2016; 8(11):320. https://0-doi-org.brum.beds.ac.uk/10.3390/v8110320
Chicago/Turabian StyleToro-Ascuy, Daniela, Bárbara Rojas-Araya, Fernando Valiente-Echeverría, and Ricardo Soto-Rifo. 2016. "Interactions between the HIV-1 Unspliced mRNA and Host mRNA Decay Machineries" Viruses 8, no. 11: 320. https://0-doi-org.brum.beds.ac.uk/10.3390/v8110320





