Seed Transmission of Beet Curly Top Virus and Beet Curly Top Iran Virus in a Local Cultivar of Petunia in Iran
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sources of Petunia Seeds and Virus Isolates
2.2. Seed Transmission Experiments
2.3. DNA Extraction, PCR Analysis and Sequencing
2.4. Immunocapture PCR
2.5. DIG Southern Blot Hybridization Analysis
2.6. Seed Transmission Rate of BCTV-Svr and BCTIV
3. Results
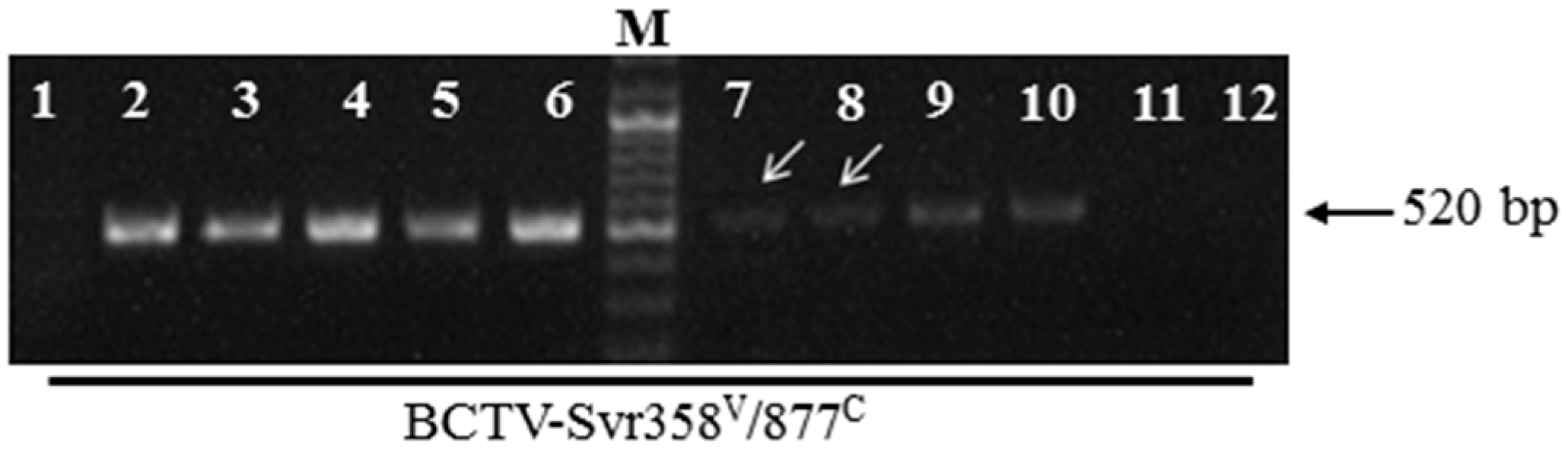
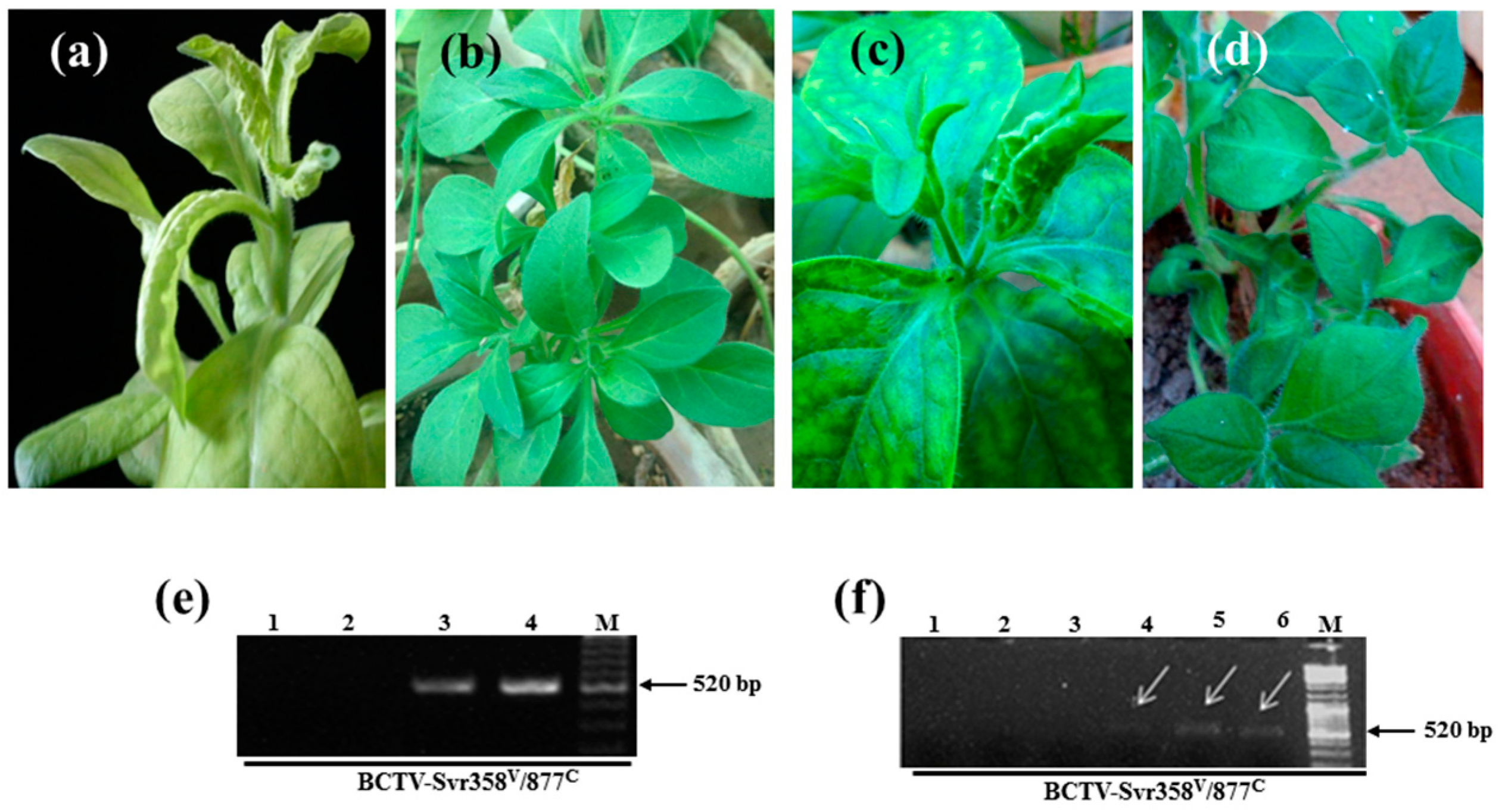
3.1. Detection of BCTV-Svr and BCTIV in Petunia Seeds and Their Developed Symptomatic Plants
3.2. Vertical Transmission of BCTV-Svr and BCTIV through Three Generations of Petunia
3.3. Presence of Virion Particles in the Seedlings and Seeds
3.4. BCTV-Svr Infection of Seeds in Agroinfected Petunia Plants
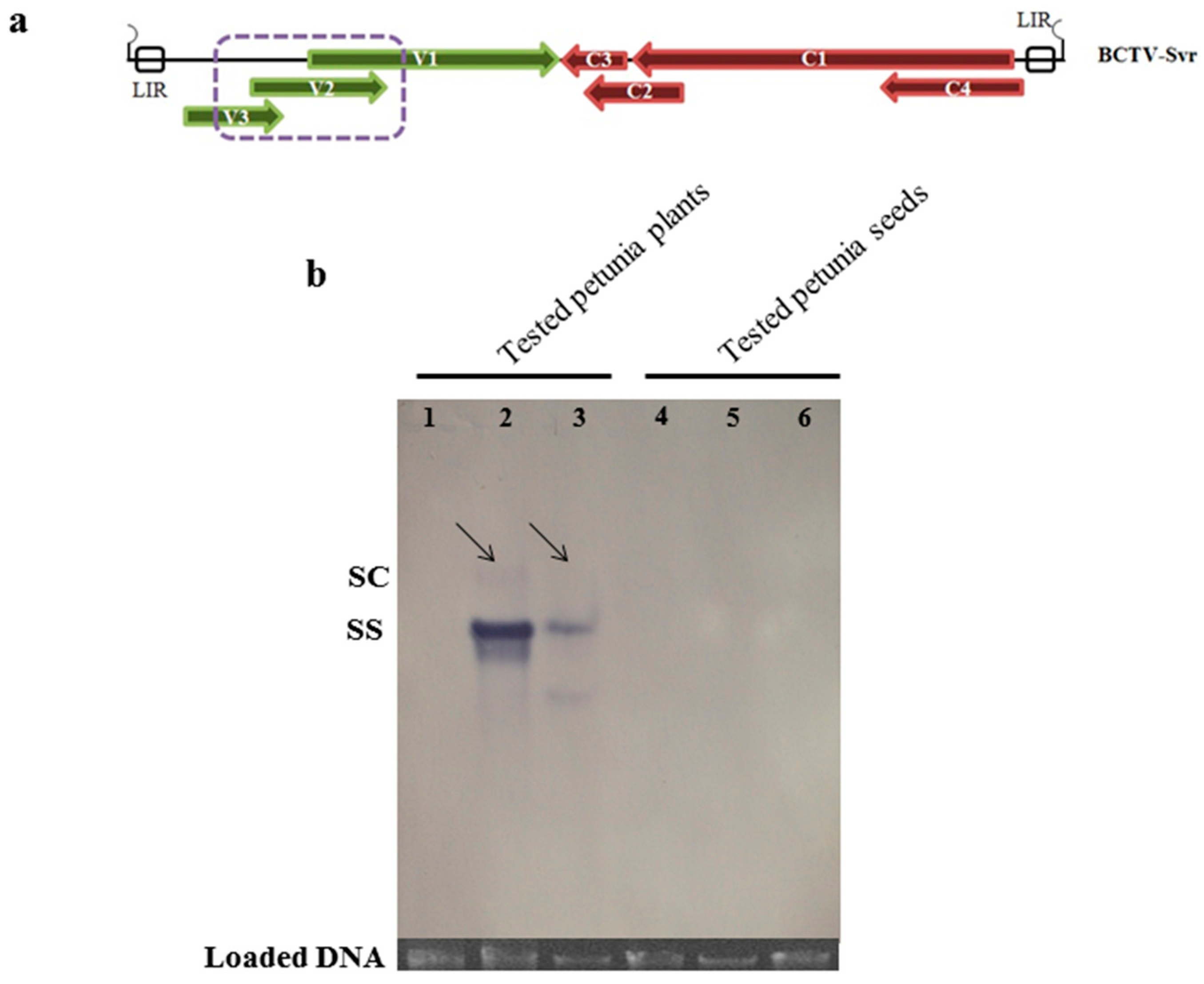
3.5. Southern Blot Analysis of BCTV-Svr-Infected Plants
3.6. Seed Transmission Rate of BCTV-Svr and BCTIV
4. Discussion
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Bennett, C.W. The Curly Top Disease of Sugar Beet and Other Plants; America Phytopathological Society: St. Paul, MN, USA, 1971. [Google Scholar]
- Gibson, K.E. Possible incidence of curly top in Iran: A new record. Plant Dis. Rep. 1967, 51, 976–977. [Google Scholar]
- Ebadzad Sahraei, G. Molecular Characterization of Iranian Isolates of Beet Curly Top Virus. Master’s Thesis, Shiraz University, Shiraz, Iran, 2008. [Google Scholar]
- Ghodoum Parizipour, M.H. Distribution of Viruses Causing Sugar Beet Curly Top Disease and the Effects of Temperature on Recovery of Beet Severe Curly Top Virus-Infected Plants. Master’s Thesis, Shiraz University, Shiraz, Iran, 2011. [Google Scholar]
- Anabestani, A.; Izadpanah, K.; Tabein, S.; Behjatnia, S.A.A. Seed transmission of beet curly top viruses in petunia. In Proceedings of the 11th Australian Plant Virology Workshop, Brisbane, Australia, 13–15 August 2014. [Google Scholar]
- Varsani, A.; Martin, D.P.; Navas-Castillo, J.; Moriones, E.; Hernandez-Zepeda, C.; Idris, A.; Zerbini, F.Z.; Brown, J.K. Revisiting the classification of curtoviruses based on genome-wide paiwise identity. Arch. Virol. 2014, 159, 1873–1882. [Google Scholar] [CrossRef] [PubMed]
- Briddon, R.W.; Stenger, D.C.; Bedford, I.D.; Stanley, J.; Izadpanah, K.; Markham, P.G. Comparisons of a beet curly top virus isolate originating from the old world with those from the new world. Eur. J. Plant Pathol. 1998, 104, 77–84. [Google Scholar] [CrossRef]
- Bolok Yazdi, H.R.; Heydarnejad, J.; Massumi, H. Genome characterization and genetic diversity of beet curly top Iran virus: A geminivirus with a novel nonanucleotide. Virus Genes 2008, 36, 539–545. [Google Scholar] [CrossRef] [PubMed]
- Soleimani, R.; Matic, S.; Taheri, H.; Behjatnia, S.A.A.; Vecchiati, M.; Izadpanah, K.; Accotto, G.P. The unconventional geminivirus Beet curly top Iran virus: Satisfying Koch’s postulates and determining vector and host range. Ann. Appl. Biol. 2013, 162, 174–181. [Google Scholar] [CrossRef]
- Taheri, H.; Izadpanah, K.; Behjatnia, S.A.A. Circulifer haematoceps, the vector of the Beet curly top Iran virus. Iran. J. Plant Pathol. 2012, 48, 45. [Google Scholar]
- Heydarnejad, J.; Keyvani, N.; Razavinejad, S.; Massumi, H.; Varsani, A. Fulfilling Koch’s postulates for beet curly top Iran virus and proposal for consideration of new genus in the family. Geminiviridae Arch. Virol. 2013, 158, 435–443. [Google Scholar] [CrossRef] [PubMed]
- Fatahi, Z.; Behjatnia, S.A.A.; Afsharifar, A.; Hamzehzarghani, H.; Izadpanah, K. Screening of sugar beet cultivars resistant to Iranian isolate of Beet severe curly top virus using an infectious clone of the virus. Iran J. Plant Pathol. 2012, 48, 111–121. [Google Scholar]
- Boulton, M.I. Agrobacterium-mediated transfer of geminiviruses to plant tissues. In Methods in Molecular Biology; Heddwyn, J., Ed.; Humana Press: Totowa, NJ, USA, 1995; Volume 49, pp. 77–93. [Google Scholar]
- Ebadzad Sahraei, G.; Behjatnia, S.A.A.; Izadpanah, K. Infectivity of the cloned genome of Iranian isolate of Beet severe curly top virus in experimental hosts. Iran J. Plant Pathol. 2008, 44, 176–183. [Google Scholar]
- Redinbaugh, M.G. Transmission of Maize streak virus by vascular puncture inoculation with unit-length genomic DNA. J. Virol. Methods 2003, 109, 95–98. [Google Scholar] [CrossRef]
- Kim, J.; Kil, E.J.; Kim, S.; Seo, H.; Byun, H.S.; Park, J.; Chung, M.N.; Kwak, H.R.; Kim, M.K.; Kim, C.S.; et al. Seed transmission of Sweet potato leaf curl virus in sweet potato (Ipomoea batatas). Plant Pathol. 2015, 64, 1284–1291. [Google Scholar] [CrossRef]
- Kil, E.J.; Kim, S.; Lee, Y.J.; Byun, H.S.; Prak, J.; Seo, H.; Kim, C.S.; Shim, J.K.; Lee, J.H.; Kim, J.K.; et al. Tomato yellow leaf curl virus (TYLCV-IL): A seed-transmissible geminivirus in tomatoes. Sci. Rep. 2016, 6, 19013. [Google Scholar] [CrossRef] [PubMed]
- Kothandaraman, S.V.; Devadason, A.; Ganesan, M.V. Seed-borne nature of a begomovirus, Mung bean yellow mosaic virus in black gram. Appl. Microbiol. Biotechnol. 2016, 100, 1925–1933. [Google Scholar] [CrossRef] [PubMed]
- Meyerdirk, D.E.; Oldfield, G.N. Monitoring Circulifer tenellus (Baker) (Homoptera: Cicadellidae). Can. Entomol. 1985, 117, 505–511. [Google Scholar] [CrossRef]
- Gawel, N.J.; Jarret, R.L. A modified CTAB DNA extraction procedure for Musa and Ipomoea. Plant Mol. Biol. Rep. 1991, 9, 262–266. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed]
- Zorriasatain, S. Host Range, Variation of the Vector Population & Purification of Beet Curly Top Virus. Master’s Thesis, Shiraz University, Shiraz, Iran, 2003. [Google Scholar]
- Tabein, S.; Behjatnia, S.A.A.; Anabestani, A.; Izadpanah, K. Whitefly-mediated transmissions of cotton leaf curl Multan betasatellite: Evidence for betasatellite encapsidation in coat protein of helper begomoviruses. Arch. Virol. 2013, 158, 19–26. [Google Scholar] [CrossRef] [PubMed]
- Behjatnia, S.A.A.; Dry, I.B.; Rezaian, M.A. Characterization and transient replication of tomato leaf curl virus defective DNAs. Arch. Virol. 2007, 152, 1127–1138. [Google Scholar] [CrossRef] [PubMed]
- Abdel-salam, A.M.; Amin, A.H. An Egyptian isolate of beet curly top virus: New differential hosts, physical properties, seed transmission, and serologic studies. Bull. Faculty Agric. Univ. Cairo 1990, 41, 843–858. [Google Scholar]
- Keur, J. Studies of the occurrence and transmission of virus diseases in the genus Abutilon. Bull. Torrey Bot. Club 1934, 6, 53–70. [Google Scholar] [CrossRef]
- Kil, E.J.; Prak, J.; Choi, H.S.; Kim, C.S.; Lee, S. Seed transmission of Tomato yellow leaf curl virus in white soybean (Glycine max). Plant Pathol. J. 2017, 33, 424–428. [Google Scholar] [PubMed]
- Rosas-Díaz, T.; Zhang, D.; Lozano-Durán, R. No evidence of seed transmissibility of Tomato yellow leaf curl virus in Nicotiana benthamiana. J. Zhejiang Univ. Sci. B 2017, 18, 437–440. [Google Scholar] [CrossRef] [PubMed]
- Hull, R. Plant Virology; Elsevier/Academic Press: London, UK, 2014. [Google Scholar]
- Maule, A.J.; Wang, D. Seed transmission of plant viruses: A lesson in biological complexity. Trends Microbiol. 1996, 4, 153–158. [Google Scholar] [PubMed]
- Hull, R. Comparative Plant Virology; Elsevier/Academic Press: London, UK, 2009. [Google Scholar]
- Sastry, K.S. Seed-Borne Plant Virus Diseases; Springer: New Dehli, India, 2013. [Google Scholar]


| Primer | * Nucleotide Position | Sequence from 5′ to 3′ | Expected Product Size (bp) |
|---|---|---|---|
| BCTV-Svr358V | 358–383 | GTGGATCAATTTCCAGACAATTATC | 520 |
| BCTV-Svr877C | 853–877 | CCCCATAAGAGCCATATCAAACTTC | |
| BCTIV474V | 474–493 | TACAAGAAGTATGGCGGTTC | 682 |
| BCTIV1155C | 1134–1155 | AAGAATAGCATTCTCCTTCAC |
| Experiment | Sampling Stage | Petunia Cultivar | Infected/Tested (Percentage of Infection) | ||
|---|---|---|---|---|---|
| BCTV-Svr | BCTIV | Mixed Infection | |||
| 1 | 4–6 leaf | local | 13/34 (38.2%) | 3/34 (8.8%) | 1/34 (2.9%) |
| PCSP | 0/23 (0%) | 0/23 (0%) | 0/23 (0%) | ||
| 2 | flowering | local | 39/50 (78.0%) | 5/50 (10.0%) | 2/50 (4.0%) |
| PCSP | 0/56 (0%) | 0/56 (0%) | 0/56 (0%) | ||
| 3 | flowering | local | 49/70 (70.0%) | 13/70 (18.5%) | 3/70 (4.2%) |
| PCSP | 0/70 (0%) | 0/70 (0%) | 0/70 (0%) | ||
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Anabestani, A.; Behjatnia, S.A.A.; Izadpanah, K.; Tabein, S.; Accotto, G.P. Seed Transmission of Beet Curly Top Virus and Beet Curly Top Iran Virus in a Local Cultivar of Petunia in Iran. Viruses 2017, 9, 299. https://0-doi-org.brum.beds.ac.uk/10.3390/v9100299
Anabestani A, Behjatnia SAA, Izadpanah K, Tabein S, Accotto GP. Seed Transmission of Beet Curly Top Virus and Beet Curly Top Iran Virus in a Local Cultivar of Petunia in Iran. Viruses. 2017; 9(10):299. https://0-doi-org.brum.beds.ac.uk/10.3390/v9100299
Chicago/Turabian StyleAnabestani, Ameneh, Seyed Ali Akbar Behjatnia, Keramat Izadpanah, Saeid Tabein, and Gian Paolo Accotto. 2017. "Seed Transmission of Beet Curly Top Virus and Beet Curly Top Iran Virus in a Local Cultivar of Petunia in Iran" Viruses 9, no. 10: 299. https://0-doi-org.brum.beds.ac.uk/10.3390/v9100299





