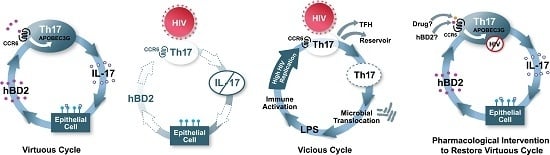
Human Beta Defensin 2 Selectively Inhibits HIV-1 in Highly Permissive CCR6+CD4+ T Cells
Abstract
:1. Introduction
2. Materials and Methods
2.1 Isolation and Culture of Primary Cells and Cell Lines
2.2. Flow Cytometry
2.3. Virus Production
2.4. Infectivity Assays
2.5. Immunoblotting
2.6. Quantitative RT-PCR
2.7. Fast Protein Liquid Chromatography
2.8. Chromatin Immunoprecipitation (ChIP) Assays
2.9. Total Chemical Synthesis of hBD2 and MIP-3α
2.10. Statistical Analysis
3. Results
3.1. CCR6+ Cells Are Protected by hBD2
3.2. hBD2 Enhances LMM and HMM APOBEC3G Expression
3.3. Induction of APOBEC3G by hBD2 Requires ERK1/2 Phosphorylation
3.4. hBD2 Induces APOBEC3G Transcription
3.5. NFAT and IRF-4 Binding after hBD2 Treatment
4. Discussion
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Lecureuil, C.; Combadiere, B.; Mazoyer, E.; Bonduelle, O.; Samri, A.; Autran, B.; Debre, P.; Combadiere, C. Trapping and apoptosis of novel subsets of memory T lymphocytes expressing CCR6 in the spleen of HIV-infected patients. Blood 2007, 109, 3649–3657. [Google Scholar] [CrossRef] [PubMed]
- Gosselin, A.; Monteiro, P.; Chomont, N.; Diaz-Griffero, F.; Said, E.A.; Fonseca, S.; Wacleche, V.; El-Far, M.; Boulassel, M.R.; Routy, J.P.; et al. Peripheral blood CCR4+CCR6+ and CXCR3+CCR6+CD4+ T cells are highly permissive to HIV-1 infection. J. Immunol. 2010, 184, 1604–1616. [Google Scholar] [CrossRef] [PubMed]
- Bouskra, D.; Brezillon, C.; Berard, M.; Werts, C.; Varona, R.; Boneca, I.G.; Eberl, G. Lymphoid tissue genesis induced by commensals through NOD1 regulates intestinal homeostasis. Nature 2008, 456, 507–510. [Google Scholar] [CrossRef] [PubMed]
- Cook, D.N.; Prosser, D.M.; Forster, R.; Zhang, J.; Kuklin, N.A.; Abbondanzo, S.J.; Niu, X.D.; Chen, S.C.; Manfra, D.J.; Wiekowski, M.T.; et al. CCR6 mediates dendritic cell localization, lymphocyte homeostasis, and immune responses in mucosal tissue. Immunity 2000, 12, 495–503. [Google Scholar] [CrossRef]
- Le Borgne, M.; Etchart, N.; Goubier, A.; Lira, S.A.; Sirard, J.C.; van Rooijen, N.; Caux, C.; Ait-Yahia, S.; Vicari, A.; Kaiserlian, D.; et al. Dendritic cells rapidly recruited into epithelial tissues via CCR6/CCL20 are responsible for CD8+ T cell crosspriming in vivo. Immunity 2006, 24, 191–201. [Google Scholar] [CrossRef] [PubMed]
- Lugering, A.; Floer, M.; Westphal, S.; Maaser, C.; Spahn, T.W.; Schmidt, M.A.; Domschke, W.; Williams, I.R.; Kucharzik, T. Absence of CCR6 inhibits CD4+ regulatory T-cell development and M-cell formation inside peyer's patches. Am. J. Pathol. 2005, 166, 1647–1654. [Google Scholar] [CrossRef]
- McDonald, K.G.; McDonough, J.S.; Wang, C.; Kucharzik, T.; Williams, I.R.; Newberry, R.D. CC chemokine receptor 6 expression by B lymphocytes is essential for the development of isolated lymphoid follicles. Am. J. Pathol. 2007, 170, 1229–1240. [Google Scholar] [CrossRef] [PubMed]
- Sallusto, F.; Lanzavecchia, A.; Mackay, C.R. Chemokines and chemokine receptors in T-cell priming and Th1/Th2-mediated responses. Immunol. Today 1998, 19, 568–574. [Google Scholar] [CrossRef]
- Williams, I.R. CCR6 and CCL20: Partners in intestinal immunity and lymphorganogenesis. Ann. N. Y. Acad. Sci. 2006, 1072, 52–61. [Google Scholar] [CrossRef] [PubMed]
- Liao, F.; Rabin, R.L.; Smith, C.S.; Sharma, G.; Nutman, T.B.; Farber, J.M. CC-chemokine receptor 6 is expressed on diverse memory subsets of T cells and determines responsiveness to macrophage inflammatory protein 3 alpha. J. Immunol. 1999, 162, 186–194. [Google Scholar] [PubMed]
- Acosta-Rodriguez, E.V.; Rivino, L.; Geginat, J.; Jarrossay, D.; Gattorno, M.; Lanzavecchia, A.; Sallusto, F.; Napolitani, G. Surface phenotype and antigenic specificity of human interleukin 17-producing T helper memory cells. Nat. Immunol. 2007, 8, 639–646. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.P.; Zhang, H.H.; Foley, J.F.; Hedrick, M.N.; Farber, J.M. Human T cells that are able to produce IL-17 express the chemokine receptor CCR6. J. Immunol. 2008, 180, 214–221. [Google Scholar] [CrossRef] [PubMed]
- El Hed, A.; Khaitan, A.; Kozhaya, L.; Manel, N.; Daskalakis, D.; Borkowsky, W.; Valentine, F.; Littman, D.R.; Unutmaz, D. Susceptibility of human Th17 cells to human immunodeficiency virus and their perturbation during infection. J. Infect. Dis. 2010, 201, 843–854. [Google Scholar] [CrossRef] [PubMed]
- Alvarez, Y.; Tuen, M.; Shen, G.; Nawaz, F.; Arthos, J.; Wolff, M.J.; Poles, M.A.; Hioe, C.E. Preferential HIV infection of CCR6+ Th17 cells is associated with higher levels of virus receptor expression and lack of CCR5 ligands. J. Virol. 2013, 87, 10843–10854. [Google Scholar] [CrossRef] [PubMed]
- Guan, Y.; Abdelwahab, S.; Kamin-Lewis, R.; DeVico, A.L.; Lewis, G.K. Self-protection of individual CD4+ T cells against R5 HIV-1 infection by the synthesis of anti-viral CCR5 ligands. PLoS ONE 2008, 3, e3481. [Google Scholar] [CrossRef] [PubMed]
- Christensen-Quick, A.; Lafferty, M.; Sun, L.; Marchionni, L.; DeVico, A.; Garzino-Demo, A. Human Th17 cells lack HIV-inhibitory rnases and are highly permissive to productive HIV infection. J. Virol. 2016, 90, 7833–7847. [Google Scholar] [CrossRef] [PubMed]
- Monteiro, P.; Gosselin, A.; Wacleche, V.S.; El-Far, M.; Said, E.A.; Kared, H.; Grandvaux, N.; Boulassel, M.R.; Routy, J.P.; Ancuta, P. Memory CCR6+CD4+ T cells are preferential targets for productive HIV type 1 infection regardless of their expression of integrin beta7. J. Immunol. 2011, 186, 4618–4630. [Google Scholar] [CrossRef] [PubMed]
- Khoury, G.; Anderson, J.L.; Fromentin, R.; Hartogenesis, W.; Smith, M.Z.; Bacchetti, P.; Hecht, F.M.; Chomont, N.; Cameron, P.U.; Deeks, S.G.; et al. Persistence of integrated HIV DNA in CXCR3+ CCR6+ memory CD4+ T cells in HIV-infected individuals on antiretroviral therapy. AIDS 2016, 30, 1511–1520. [Google Scholar] [CrossRef] [PubMed]
- Wacleche, V.S.; Goulet, J.P.; Gosselin, A.; Monteiro, P.; Soudeyns, H.; Fromentin, R.; Jenabian, M.A.; Vartanian, S.; Deeks, S.G.; Chomont, N.; et al. New insights into the heterogeneity of Th17 subsets contributing to HIV-1 persistence during antiretroviral therapy. Retrovirology 2016, 13, 59. [Google Scholar] [CrossRef] [PubMed]
- Gosselin, A.; Wiche Salinas, T.R.; Planas, D.; Wacleche, V.S.; Zhang, Y.; Fromentin, R.; Chomont, N.; Cohen, E.A.; Shacklett, B.; Mehraj, V.; et al. HIV persists in CCR6+CD4+ T cells from colon and blood during antiretroviral therapy. AIDS 2017, 31, 35–48. [Google Scholar] [CrossRef] [PubMed]
- Brenchley, J.M.; Paiardini, M.; Knox, K.S.; Asher, A.I.; Cervasi, B.; Asher, T.E.; Scheinberg, P.; Price, D.A.; Hage, C.A.; Kholi, L.M.; et al. Differential Th17 CD4 T-cell depletion in pathogenic and nonpathogenic lentiviral infections. Blood 2008, 112, 2826–2835. [Google Scholar] [CrossRef] [PubMed]
- Cecchinato, V.; Trindade, C.J.; Laurence, A.; Heraud, J.M.; Brenchley, J.M.; Ferrari, M.G.; Zaffiri, L.; Tryniszewska, E.; Tsai, W.P.; Vaccari, M.; et al. Altered balance between Th17 and Th1 cells at mucosal sites predicts aids progression in simian immunodeficiency virus-infected macaques. Mucosal Immunol. 2008, 1, 279–288. [Google Scholar] [CrossRef] [PubMed]
- Stieh, D.J.; Matias, E.; Xu, H.; Fought, A.J.; Blanchard, J.L.; Marx, P.A.; Veazey, R.S.; Hope, T.J. Th17 cells are preferentially infected very early after vaginal transmission of SIV in macaques. Cell Host Microbe 2016, 19, 529–540. [Google Scholar] [CrossRef] [PubMed]
- McGary, C.S.; Alvarez, X.; Harrington, S.; Cervasi, B.; Ryan, E.S.; Iriele, R.I.; Paganini, S.; Harper, J.L.; Easley, K.; Silvestri, G.; et al. The loss of CCR6+ and CD161+ CD4+ T-cell homeostasis contributes to disease progression in SIV-infected rhesus macaques. Mucosal Immunol. 2017. [Google Scholar] [CrossRef] [PubMed]
- Brenchley, J.M.; Schacker, T.W.; Ruff, L.E.; Price, D.A.; Taylor, J.H.; Beilman, G.J.; Nguyen, P.L.; Khoruts, A.; Larson, M.; Haase, A.T.; et al. Cd4+ T cell depletion during all stages of HIV disease occurs predominantly in the gastrointestinal tract. J. Exp. Med. 2004, 200, 749–759. [Google Scholar] [CrossRef] [PubMed]
- Mehandru, S.; Poles, M.A.; Tenner-Racz, K.; Horowitz, A.; Hurley, A.; Hogan, C.; Boden, D.; Racz, P.; Markowitz, M. Primary HIV-1 infection is associated with preferential depletion of CD4+ T lymphocytes from effector sites in the gastrointestinal tract. J. Exp. Med. 2004, 200, 761–770. [Google Scholar] [CrossRef] [PubMed]
- Mehandru, S.; Poles, M.A.; Tenner-Racz, K.; Jean-Pierre, P.; Manuelli, V.; Lopez, P.; Shet, A.; Low, A.; Mohri, H.; Boden, D.; et al. Lack of mucosal immune reconstitution during prolonged treatment of acute and early HIV-1 infection. PLoS Med. 2006, 3, e484. [Google Scholar] [CrossRef]
- Brenchley, J.M.; Price, D.A.; Schacker, T.W.; Asher, T.E.; Silvestri, G.; Rao, S.; Kazzaz, Z.; Bornstein, E.; Lambotte, O.; Altmann, D.; et al. Microbial translocation is a cause of systemic immune activation in chronic HIV infection. Nat. Med. 2006, 12, 1365–1371. [Google Scholar] [CrossRef] [PubMed]
- Raffatellu, M.; Santos, R.L.; Verhoeven, D.E.; George, M.D.; Wilson, R.P.; Winter, S.E.; Godinez, I.; Sankaran, S.; Paixao, T.A.; Gordon, M.A.; et al. Simian immunodeficiency virus-induced mucosal interleukin-17 deficiency promotes salmonella dissemination from the gut. Nat. Med. 2008, 14, 421–428. [Google Scholar] [CrossRef] [PubMed]
- Ciccone, E.J.; Greenwald, J.H.; Lee, P.I.; Biancotto, A.; Read, S.W.; Yao, M.A.; Hodge, J.N.; Thompson, W.L.; Kovacs, S.B.; Chairez, C.L.; et al. CD4+ T cells, including Th17 and cycling subsets, are intact in the gut mucosa of HIV-1-infected long-term nonprogressors. J. Virol. 2011, 85, 5880–5888. [Google Scholar] [CrossRef] [PubMed]
- Chege, D.; Sheth, P.M.; Kain, T.; Kim, C.J.; Kovacs, C.; Loutfy, M.; Halpenny, R.; Kandel, G.; Chun, T.W.; Ostrowski, M.; et al. Sigmoid Th17 populations, the HIV latent reservoir, and microbial translocation in men on long-term antiretroviral therapy. AIDS 2011, 25, 741–749. [Google Scholar] [CrossRef] [PubMed]
- Curtis, M.M.; Way, S.S. Interleukin-17 in host defence against bacterial, mycobacterial and fungal pathogens. Immunology 2009, 126, 177–185. [Google Scholar] [CrossRef] [PubMed]
- Conti, H.R.; Shen, F.; Nayyar, N.; Stocum, E.; Sun, J.N.; Lindemann, M.J.; Ho, A.W.; Hai, J.H.; Yu, J.J.; Jung, J.W.; et al. Th17 cells and IL-17 receptor signaling are essential for mucosal host defense against oral candidiasis. J. Exp. Med. 2009, 206, 299–311. [Google Scholar] [CrossRef] [PubMed]
- Klatt, N.R.; Brenchley, J.M. Th17 cell dynamics in HIV infection. Curr. Opin. HIV AIDS 2010, 5, 135–140. [Google Scholar] [CrossRef] [PubMed]
- Hernandez-Santos, N.; Huppler, A.R.; Peterson, A.C.; Khader, S.A.; McKenna, K.C.; Gaffen, S.L. Th17 cells confer long-term adaptive immunity to oral mucosal Candida albicans infections. Mucosal Immunol. 2013, 6, 900–910. [Google Scholar] [CrossRef] [PubMed]
- Fossiez, F.; Djossou, O.; Chomarat, P.; Flores-Romo, L.; Ait-Yahia, S.; Maat, C.; Pin, J.J.; Garrone, P.; Garcia, E.; Saeland, S.; et al. T cell interleukin-17 induces stromal cells to produce proinflammatory and hematopoietic cytokines. J. Exp. Med. 1996, 183, 2593–2603. [Google Scholar] [CrossRef] [PubMed]
- LeGrand, A.; Fermor, B.; Fink, C.; Pisetsky, D.S.; Weinberg, J.B.; Vail, T.P.; Guilak, F. Interleukin-1, tumor necrosis factor alpha, and interleukin-17 synergistically up-regulate nitric oxide and prostaglandin E2 production in explants of human osteoarthritic knee menisci. Arthritis Rheum. 2001, 44, 2078–2083. [Google Scholar] [CrossRef]
- Moseley, T.A.; Haudenschild, D.R.; Rose, L.; Reddi, A.H. Interleukin-17 family and IL-17 receptors. Cytokine Growth Factor Rev. 2003, 14, 155–174. [Google Scholar] [CrossRef]
- Ye, P.; Rodriguez, F.H.; Kanaly, S.; Stocking, K.L.; Schurr, J.; Schwarzenberger, P.; Oliver, P.; Huang, W.; Zhang, P.; Zhang, J.; et al. Requirement of interleukin 17 receptor signaling for lung CXC chemokine and granulocyte colony-stimulating factor expression, neutrophil recruitment, and host defense. J. Exp. Med. 2001, 194, 519–527. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Thai, P.; Zhao, Y.H.; Ho, Y.S.; DeSouza, M.M.; Wu, R. Stimulation of airway mucin gene expression by interleukin (IL)-17 through IL-6 paracrine/autocrine loop. J. Biol. Chem. 2003, 278, 17036–17043. [Google Scholar] [CrossRef] [PubMed]
- Kao, C.Y.; Chen, Y.; Thai, P.; Wachi, S.; Huang, F.; Kim, C.; Harper, R.W.; Wu, R. IL-17 markedly up-regulates beta-defensin-2 expression in human airway epithelium via JAK and NF-kappaB signaling pathways. J. Immunol. 2004, 173, 3482–3491. [Google Scholar] [CrossRef] [PubMed]
- Kao, C.Y.; Huang, F.; Chen, Y.; Thai, P.; Wachi, S.; Kim, C.; Tam, L.; Wu, R. Up-regulation of CC chemokine ligand 20 expression in human airway epithelium by IL-17 through a JAK-independent but MEK/NF-KappaB-dependent signaling pathway. J. Immunol. 2005, 175, 6676–6685. [Google Scholar] [CrossRef] [PubMed]
- Kinugasa, T.; Sakaguchi, T.; Gu, X.; Reinecker, H.C. Claudins regulate the intestinal barrier in response to immune mediators. Gastroenterology 2000, 118, 1001–1011. [Google Scholar] [CrossRef]
- Liang, S.C.; Tan, X.Y.; Luxenberg, D.P.; Karim, R.; Dunussi-Joannopoulos, K.; Collins, M.; Fouser, L.A. Interleukin (IL)-22 and IL-17 are coexpressed by Th17 cells and cooperatively enhance expression of antimicrobial peptides. J. Exp. Med. 2006, 203, 2271–2279. [Google Scholar] [CrossRef] [PubMed]
- Wolk, K.; Kunz, S.; Witte, E.; Friedrich, M.; Asadullah, K.; Sabat, R. IL-22 increases the innate immunity of tissues. Immunity 2004, 21, 241–254. [Google Scholar] [CrossRef] [PubMed]
- Lehrer, R.I.; Ganz, T.; Selsted, M.E. Defensins: Endogenous antibiotic peptides of animal cells. Cell 1991, 64, 229–230. [Google Scholar] [CrossRef]
- Zasloff, M. Antibiotic peptides as mediators of innate immunity. Curr. Opin. Immunol. 1992, 4, 3–7. [Google Scholar] [CrossRef]
- Ganz, T. Defensins: Antimicrobial peptides of innate immunity. Nat. Rev. Immunol. 2003, 3, 710–720. [Google Scholar] [CrossRef] [PubMed]
- Selsted, M.E.; Ouellette, A.J. Mammalian defensins in the antimicrobial immune response. Nat. Immunol. 2005, 6, 551–557. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Xu, Z.; Peng, L.; Fang, X.; Yin, X.; Xu, N.; Cen, P. Recent advances in the research and development of human defensins. Peptides 2006, 27, 931–940. [Google Scholar] [CrossRef] [PubMed]
- Pazgier, M.; Hoover, D.M.; Yang, D.; Lu, W.; Lubkowski, J. Human beta-defensins. Cell. Mol. Life Sci. 2006, 63, 1294–1313. [Google Scholar] [CrossRef] [PubMed]
- Ding, J.; Chou, Y.Y.; Chang, T.L. Defensins in viral infections. J. Innate Immun. 2009, 1, 413–420. [Google Scholar] [CrossRef] [PubMed]
- Wilson, S.S.; Wiens, M.E.; Smith, J.G. Antiviral mechanisms of human defensins. J. Mol. Biol. 2013, 425, 4965–4980. [Google Scholar] [CrossRef] [PubMed]
- Bevins, C.L. Innate immune functions of alpha-defensins in the small intestine. Dig. Dis. 2013, 31, 299–304. [Google Scholar] [CrossRef] [PubMed]
- Daher, K.A.; Selsted, M.E.; Lehrer, R.I. Direct inactivation of viruses by human granulocyte defensins. J. Virol. 1986, 60, 1068–1074. [Google Scholar] [PubMed]
- Chang, T.L.; Klotman, M.E. Defensins: Natural anti-HIV peptides. AIDS Rev. 2004, 6, 161–168. [Google Scholar] [PubMed]
- Garzino-Demo, A. Chemokines and defensins as HIV suppressive factors: An evolving story. Curr. Pharm. Des. 2007, 13, 163–172. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.; Chertov, O.; Bykovskaia, S.N.; Chen, Q.; Buffo, M.J.; Shogan, J.; Anderson, M.; Schroder, J.M.; Wang, J.M.; Howard, O.M.; et al. Beta-defensins: Linking innate and adaptive immunity through dendritic and T cell CCR6. Science 1999, 286, 525–528. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Hoover, D.M.; Yang, D.; Boulegue, C.; Santamaria, F.; Oppenheim, J.J.; Lubkowski, J.; Lu, W. Engineering disulfide bridges to dissect antimicrobial and chemotactic activities of human beta-defensin 3. Proc. Natl. Acad. Sci. USA 2003, 100, 8880–8885. [Google Scholar] [CrossRef] [PubMed]
- Niyonsaba, F.; Ogawa, H.; Nagaoka, I. Human beta-defensin-2 functions as a chemotactic agent for tumour necrosis factor-alpha-treated human neutrophils. Immunology 2004, 111, 273–281. [Google Scholar] [CrossRef] [PubMed]
- Soruri, A.; Grigat, J.; Forssmann, U.; Riggert, J.; Zwirner, J. Beta-defensins chemoattract macrophages and mast cells but not lymphocytes and dendritic cells: CCR6 is not involved. Eur. J. Immunol. 2007, 37, 2474–2486. [Google Scholar] [CrossRef] [PubMed]
- Barabas, N.; Rohrl, J.; Holler, E.; Hehlgans, T. Beta-defensins activate macrophages and synergize in pro-inflammatory cytokine expression induced by TLR ligands. Immunobiology 2013, 218, 1005–1011. [Google Scholar] [CrossRef] [PubMed]
- Rohrl, J.; Yang, D.; Oppenheim, J.J.; Hehlgans, T. Human beta-defensin 2 and 3 and their mouse orthologs induce chemotaxis through interaction with CCR2. J. Immunol. 2010, 184, 6688–6694. [Google Scholar] [CrossRef] [PubMed]
- Boniotto, M.; Jordan, W.J.; Eskdale, J.; Tossi, A.; Antcheva, N.; Crovella, S.; Connell, N.D.; Gallagher, G. Human beta-defensin 2 induces a vigorous cytokine response in peripheral blood mononuclear cells. Antimicrob. Agents Chemother. 2006, 50, 1433–1441. [Google Scholar] [CrossRef] [PubMed]
- Niyonsaba, F.; Ushio, H.; Nakano, N.; Ng, W.; Sayama, K.; Hashimoto, K.; Nagaoka, I.; Okumura, K.; Ogawa, H. Antimicrobial peptides human beta-defensins stimulate epidermal keratinocyte migration, proliferation and production of proinflammatory cytokines and chemokines. J. Investig. Dermatol. 2007, 127, 594–604. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Niyonsaba, F.; Ushio, H.; Hara, M.; Yokoi, H.; Matsumoto, K.; Saito, H.; Nagaoka, I.; Ikeda, S.; Okumura, K.; et al. Antimicrobial peptides human beta-defensin (HBD)-3 and HBD-4 activate mast cells and increase skin vascular permeability. Eur. J. Immunol. 2007, 37, 434–444. [Google Scholar] [CrossRef] [PubMed]
- Otte, J.M.; Werner, I.; Brand, S.; Chromik, A.M.; Schmitz, F.; Kleine, M.; Schmidt, W.E. Human beta defensin 2 promotes intestinal wound healing in vitro. J. Cell. Biochem. 2008, 104, 2286–2297. [Google Scholar] [CrossRef] [PubMed]
- Weinberg, A.; Jin, G.; Sieg, S.; McCormick, T.S. The yin and yang of human beta-defensins in health and disease. Front. Immunol. 2012, 3, 294. [Google Scholar] [CrossRef] [PubMed]
- Quinones-Mateu, M.E.; Lederman, M.M.; Feng, Z.; Chakraborty, B.; Weber, J.; Rangel, H.R.; Marotta, M.L.; Mirza, M.; Jiang, B.; Kiser, P.; et al. Human epithelial beta-defensins 2 and 3 inhibit HIV-1 replication. AIDS 2003, 17, F39–F48. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Finnegan, C.M.; Kish-Catalone, T.; Blumenthal, R.; Garzino-Demo, P.; La Terra Maggiore, G.M.; Berrone, S.; Kleinman, C.; Wu, Z.; Abdelwahab, S.; et al. Human beta-defensins suppress human immunodeficiency virus infection: Potential role in mucosal protection. J. Virol. 2005, 79, 14318–14329. [Google Scholar] [CrossRef] [PubMed]
- Lafferty, M.K.; Sun, L.; DeMasi, L.; Lu, W.; Garzino-Demo, A. CCR6 ligands inhibit HIV by inducing APOBEC3G. Blood 2010, 115, 1564–1571. [Google Scholar] [CrossRef] [PubMed]
- Sheehy, A.M.; Gaddis, N.C.; Choi, J.D.; Malim, M.H. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature 2002, 418, 646–650. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Yang, B.; Pomerantz, R.J.; Zhang, C.; Arunachalam, S.C.; Gao, L. The cytidine deaminase CEM15 induces hypermutation in newly synthesized HIV-1 DNA. Nature 2003, 424, 94–98. [Google Scholar] [CrossRef] [PubMed]
- Bishop, K.N.; Holmes, R.K.; Malim, M.H. Antiviral potency of APOBEC proteins does not correlate with cytidine deamination. J. Virol. 2006, 80, 8450–8458. [Google Scholar] [CrossRef] [PubMed]
- Bishop, K.N.; Verma, M.; Kim, E.Y.; Wolinsky, S.M.; Malim, M.H. APOBEC3G inhibits elongation of HIV-1 reverse transcripts. PLoS Pathog. 2008, 4, e1000231. [Google Scholar] [CrossRef] [PubMed]
- De Pasquale, M.; Kourteva, Y.; Allos, T.; D’Aquila, R.T. Lower HIV provirus levels are associated with more APOBEC3G protein in blood resting memory CD4+ T lymphocytes of controllers in vivo. PLoS ONE 2013, 8, e76002. [Google Scholar] [CrossRef]
- Kourteva, Y.; De Pasquale, M.; Allos, T.; McMunn, C.; D’Aquila, R.T. APOBEC3G expression and hypermutation are inversely associated with human immunodeficiency virus type 1 (HIV-1) burden in vivo. Virology 2012, 430, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Vetter, M.L.; D’Aquila, R.T. Cytoplasmic APOBEC3G restricts incoming Vif-positive human immunodeficiency virus type 1 and increases two-long terminal repeat circle formation in activated T-helper-subtype cells. J. Virol. 2009, 83, 8646–8654. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Chiu, Y.L.; Witkowska, H.E.; Hall, S.C.; Santiago, M.; Soros, V.B.; Esnault, C.; Heidmann, T.; Greene, W.C. High-molecular-mass APOBEC3G complexes restrict alu retrotransposition. Proc. Natl. Acad. Sci. USA 2006, 103, 15588–15593. [Google Scholar] [CrossRef] [PubMed]
- Opi, S.; Takeuchi, H.; Kao, S.; Khan, M.A.; Miyagi, E.; Goila-Gaur, R.; Iwatani, Y.; Levin, J.G.; Strebel, K. Monomeric APOBEC3G is catalytically active and has antiviral activity. J. Virol. 2006, 80, 4673–4682. [Google Scholar] [CrossRef] [PubMed]
- Goila-Gaur, R.; Khan, M.A.; Miyagi, E.; Kao, S.; Opi, S.; Takeuchi, H.; Strebel, K. HIV-1 Vif promotes the formation of high molecular mass APOBEC3G complexes. Virology 2008, 372, 136–146. [Google Scholar] [CrossRef] [PubMed]
- Shultz, L.D.; Saito, Y.; Najima, Y.; Tanaka, S.; Ochi, T.; Tomizawa, M.; Doi, T.; Sone, A.; Suzuki, N.; Fujiwara, H.; et al. Generation of functional human T-cell subsets with hla-restricted immune responses in HLA class I expressing NOD/SCID/IL2r gamma(null) humanized mice. Proc. Natl. Acad. Sci. USA 2010, 107, 13022–13027. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.; Huang, J.; Zhang, C.; Huang, S.; Nunnari, G.; Wang, F.X.; Tong, X.; Gao, L.; Nikisher, K.; Zhang, H. Alpha interferon potently enhances the anti-human immunodeficiency virus type 1 activity of APOBEC3G in resting primary CD4 T cells. J. Virol. 2006, 80, 7645–7657. [Google Scholar] [CrossRef] [PubMed]
- Rose, K.M.; Marin, M.; Kozak, S.L.; Kabat, D. Transcriptional regulation of APOBEC3G, a cytidine deaminase that hypermutates human immunodeficiency virus. J. Biol. Chem. 2004, 279, 41744–41749. [Google Scholar] [CrossRef] [PubMed]
- Stopak, K.S.; Chiu, Y.L.; Kropp, J.; Grant, R.M.; Greene, W.C. Distinct patterns of cytokine regulation of APOBEC3G expression and activity in primary lymphocytes, macrophages, and dendritic cells. J. Biol. Chem. 2007, 282, 3539–3546. [Google Scholar] [CrossRef] [PubMed]
- Lin, A.; Minden, A.; Martinetto, H.; Claret, F.X.; Lange-Carter, C.; Mercurio, F.; Johnson, G.L.; Karin, M. Identification of a dual specificity kinase that activates the jun kinases and p38-MPK2. Science 1995, 268, 286–290. [Google Scholar] [CrossRef] [PubMed]
- Farrow, M.A.; Kim, E.Y.; Wolinsky, S.M.; Sheehy, A.M. NFAT and IRF proteins regulate transcription of the anti-HIV protein, APOBEC3G. J. Biol. Chem. 2011, 286, 2567–2577. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Estes, J.D.; Schlievert, P.M.; Duan, L.; Brosnahan, A.J.; Southern, P.J.; Reilly, C.S.; Peterson, M.L.; Schultz-Darken, N.; Brunner, K.G.; et al. Glycerol monolaurate prevents mucosal SIV transmission. Nature 2009, 458, 1034–1038. [Google Scholar] [CrossRef] [PubMed]
- Guillot-Delost, M.; Le Gouvello, S.; Mesel-Lemoine, M.; Cherai, M.; Baillou, C.; Simon, A.; Levy, Y.; Weiss, L.; Louafi, S.; Chaput, N.; et al. Human CD90 identifies Th17/Tc17 T cell subsets that are depleted in HIV-infected patients. J. Immunol. 2011, 188, 981–991. [Google Scholar] [CrossRef] [PubMed]
- Shen, F.; Ruddy, M.J.; Plamondon, P.; Gaffen, S.L. Cytokines link osteoblasts and inflammation: Microarray analysis of interleukin-17- and TNF-alpha-induced genes in bone cells. J. Leukoc. Biol. 2005, 77, 388–399. [Google Scholar] [CrossRef] [PubMed]
- Ruddy, M.J.; Shen, F.; Smith, J.B.; Sharma, A.; Gaffen, S.L. Interleukin-17 regulates expression of the CXC chemokine LIX/CXCL5 in osteoblasts: Implications for inflammation and neutrophil recruitment. J. Leukoc. Biol. 2004, 76, 135–144. [Google Scholar] [CrossRef] [PubMed]
- Hirota, K.; Turner, J.E.; Villa, M.; Duarte, J.H.; Demengeot, J.; Steinmetz, O.M.; Stockinger, B. Plasticity of Th17 cells in peyer’s patches is responsible for the induction of T cell-dependent IgA responses. Nat. Immunol. 2013, 14, 372–379. [Google Scholar] [CrossRef] [PubMed]
- Lindqvist, M.; van Lunzen, J.; Soghoian, D.Z.; Kuhl, B.D.; Ranasinghe, S.; Kranias, G.; Flanders, M.D.; Cutler, S.; Yudanin, N.; Muller, M.I.; et al. Expansion of HIV-specific T follicular helper cells in chronic HIV infection. J. Clin. Investig. 2012, 122, 3271–3280. [Google Scholar] [CrossRef] [PubMed]
- Petrovas, C.; Yamamoto, T.; Gerner, M.Y.; Boswell, K.L.; Wloka, K.; Smith, E.C.; Ambrozak, D.R.; Sandler, N.G.; Timmer, K.J.; Sun, X.; et al. CD4 T follicular helper cell dynamics during SIV infection. J. Clin. Investig. 2012, 122, 3281–3294. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Weatherall, C.; Bailey, M.; Alcantara, S.; De Rose, R.; Estaquier, J.; Wilson, K.; Suzuki, K.; Corbeil, J.; Cooper, D.A.; et al. Simian immunodeficiency virus infects follicular helper CD4 T cells in lymphoid tissues during pathogenic infection of pigtail macaques. J. Virol. 2013, 87, 3760–3773. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Fan, X.; Auclair, S.; Ferguson, M.; Sun, J.; Soong, L.; Hou, W.; Redfield, R.R.; Birx, D.L.; Ratto-Kim, S.; et al. Sequential dysfunction and progressive depletion of candida albicans-specific CD4 T cell response in HIV-1 infection. PLoS Pathog. 2016, 12, e1005663. [Google Scholar] [CrossRef] [PubMed]
- Neidleman, J.A.; Chen, J.C.; Kohgadai, N.; Muller, J.A.; Laustsen, A.; Thavachelvam, K.; Jang, K.S.; Sturzel, C.M.; Jones, J.J.; Ochsenbauer, C.; et al. Mucosal stromal fibroblasts markedly enhance HIV infection of CD4+ T cells. PLoS Pathog. 2017, 13, e1006163. [Google Scholar] [CrossRef] [PubMed]






© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lafferty, M.K.; Sun, L.; Christensen-Quick, A.; Lu, W.; Garzino-Demo, A. Human Beta Defensin 2 Selectively Inhibits HIV-1 in Highly Permissive CCR6+CD4+ T Cells. Viruses 2017, 9, 111. https://0-doi-org.brum.beds.ac.uk/10.3390/v9050111
Lafferty MK, Sun L, Christensen-Quick A, Lu W, Garzino-Demo A. Human Beta Defensin 2 Selectively Inhibits HIV-1 in Highly Permissive CCR6+CD4+ T Cells. Viruses. 2017; 9(5):111. https://0-doi-org.brum.beds.ac.uk/10.3390/v9050111
Chicago/Turabian StyleLafferty, Mark K., Lingling Sun, Aaron Christensen-Quick, Wuyuan Lu, and Alfredo Garzino-Demo. 2017. "Human Beta Defensin 2 Selectively Inhibits HIV-1 in Highly Permissive CCR6+CD4+ T Cells" Viruses 9, no. 5: 111. https://0-doi-org.brum.beds.ac.uk/10.3390/v9050111