Geminiviruses and Plant Hosts: A Closer Examination of the Molecular Arms Race
Abstract
:1. Introduction
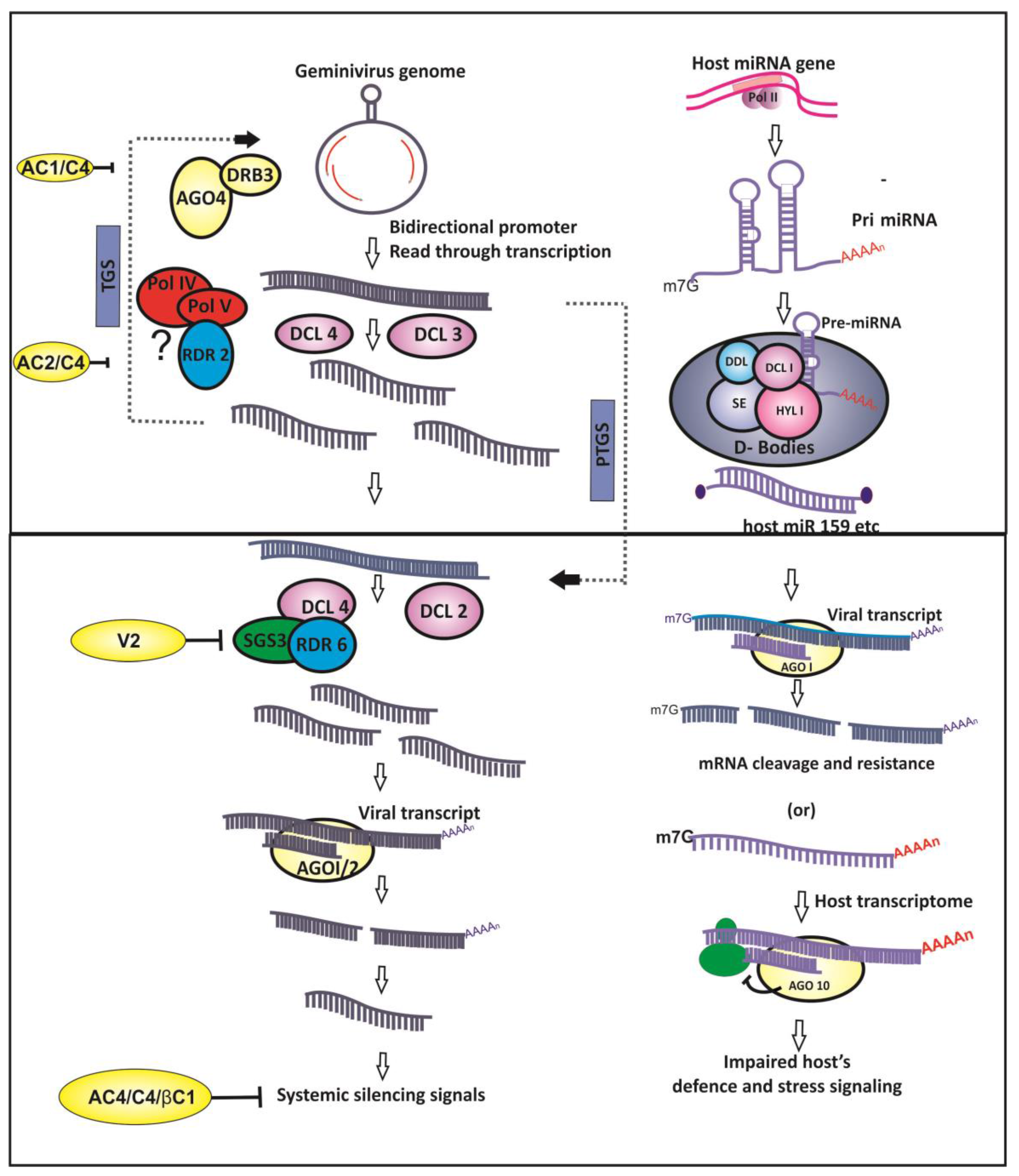
2. Geminiviruses and RNA Silencing
3. Geminivirus and Small Non-Coding RNAs
4. Geminivirus and Counter-RNA Silencing
5. Geminiviruses and Host Cellular Reprogramming
5.1. Redirection of Host Gene Expression
5.2. Host Hormonal Signaling
5.3. Altered Host Protein Degradation Pathways
5.4. Impaired Cellular Metabolism
6. Implications for Engineered Resistance
7. Future Directions and Conclusions
Author Contributions
Conflicts of Interest
Abbreviations
| ADK | adenosine kinase |
| AGO | Argonaute |
| Cas9 | CRISPR-associated 9 |
| CRISPRs | Clustered regularly interspaced short palindromic repeats |
| DCL | Dicer-like enzyme |
| HEN | HUA Enhancer |
| miRNA | MicroRNA |
| NLS | Nuclear localization signal |
| PTGS | Post-transcriptional gene silencing |
| PTM | Post translational modification |
| RdDM | RNA-dependent DNA methylation |
| RdRP | RNA-dependent RNA polymerase |
| RecSat | Recombinant satellite molecule |
| Rep | Replication initiator protein |
| RISC | RNA-induced silencing complex |
| RITS | RNA-induced transcriptional silencing |
| SAM | S-adenosyl methionine |
| SAMDC | S-adenosyl methionine decarboxylase |
| siRNA | Small interfering RNA |
| sncRNAs | Small non-coding RNAs |
| SnRK | Serine/threonine kinase (SNF) related kinase |
| TGS | Transcriptional gene silencing |
| vsiRNA | Viral genome derived small interfering RNA |
| VSR | Viral suppressors of RNA silencing |
References
- Brown, J.K.; Fauquet, C.M.; Briddon, R.W.; Zerbini, M.; Moriones, E.; Navas-Castillo, J. Family Geminiviridae. In Virus Taxonomy. Classification and Nomenclature of Viruses; Ninth Report of the International Committee on Taxonomy of Viruses; King, A.M.Q., Adams, M.J., Carstens, E.B., Lefkowitz, E.J., Eds.; Elsevier Academic Press: London, UK, 2012; pp. 351–373. [Google Scholar]
- Varsani, A.; Roumagnac, P.; Fuchs, M.; Navas-Castillo, J.; Moriones, E.; Idris, A.; Briddon, R.W.; Rivera-Bustamante, R.; Zerbini, F.M.; Martin, D.P. Capulavirus and Grablovirus: Two new genera in the family Geminiviridae. Arch. Virol. 2017, 162, 1819–1831. [Google Scholar] [CrossRef] [PubMed]
- Accotto, G.P.; Mullineaux, P.M.; Brown, S.C.; Marie, D. Digitaria streak geminivirus replicative forms are abundant in S-phase nuclei of infected cells. Virology 1993, 195, 257–259. [Google Scholar] [CrossRef] [PubMed]
- Choi, I.R.; Stenger, D.C. Strain-specific determinants of beet curly top geminivirus DNA replication. Virology 1995, 206, 904–912. [Google Scholar] [CrossRef] [PubMed]
- Bisaro, D.M. Geminivirus replication. In DNA Replication in Eukaryotic Cells; De Pamphilis, M., Ed.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 1996; pp. 833–854. [Google Scholar]
- Bisaro, D.M.; Hamilton, W.D.O.; Coutts, R.H.A.; Buck, K.W. Molecular cloning and characterization of the two DNA components of tomato golden mosaic virus. Nucleic Acids Res. 1982, 10, 4913–4922. [Google Scholar] [CrossRef] [PubMed]
- Harrison, B.D.; Robinson, D.J. Natural Genomic and antigenic variation in whitefly transmitted geminiviruses (Begomoviruses). Annu. Rev. Phytopathol. 1999, 37, 369–398. [Google Scholar] [CrossRef] [PubMed]
- Briddon, R.W.; Patil, B.L.; Bagewadi, B.; Nawaz-ul Rehman, M.S.; Fauquet, C.M. Distinct evolutionary histories of the DNAA and DNAB components of bipartite begomoviruses. BMC Evol. Biol. 2000, 10, 97. [Google Scholar]
- Briddon, R.W.; Pinner, M.S.; Stanley, J.; Markham, P.G. Geminivirus coat protein gene replacement alters insect specificity. Virology 1990, 177, 85–94. [Google Scholar] [CrossRef]
- Hanley-Bowdoin, L.; Settlage, S.B.; Orozco, B.M.; Nagar, S.; Robertson, D. Geminiviruses: Models for plant DNA replication, transcription, and cell cycle regulation. Crit. Rev. Biochem. Mol.Biol. 2000, 35, 105–140. [Google Scholar] [CrossRef] [PubMed]
- Priyadarshini, C.G.P.; Ambika, M.V.; Tippeswamy, R.; Savithri, H.S. Functional characterization of coat protein and V2 involved in cell to cell movement of Cotton leaf curlKokhran virus-Dabawali. PLoS ONE 2011, 6, e26929. [Google Scholar]
- Yang, X.; Xie, Y.; Raja, P.; Li, S.; Wolf, J.N.; Shen, Q.; Bisaro, D.M.; Zhou, X. Suppression of methylation-Mediated transcriptional gene silencing by βC1-SAHH protein interaction during Geminivirus-Betasatellite infection. PLoS Pathog. 2011, 7, e1002329. [Google Scholar] [CrossRef] [PubMed]
- Briddon, R.W.; Bull, S.E.; Amin, I.; Mansoor, S.; Bedford, I.D.; Rishi, N.; Siwatch, S.S.; Zafar, Y.; Abdel-salam, A.M.; Markham, P.G. Diversity of DNA 1: A satellite-like molecule associated with monopartite begomovirus-DNA β complexes. Virology 2004, 324, 462–474. [Google Scholar] [CrossRef] [PubMed]
- Briddon, R.W.; Stanley, J. Subviral agents associated with plant single-stranded DNA viruses. Virology 2006, 344, 198–210. [Google Scholar] [CrossRef] [PubMed]
- Saunders, K.; Bedford, I.D.; Briddon, R.W.; Markham, P.G.; Wong, S.M.; Stanley, J. A unique virus complex causes Ageratum yellow vein disease. Proc. Natl. Acad. Sci. USA 2000, 97, 6890–6895. [Google Scholar] [CrossRef] [PubMed]
- Briddon, R.W.; Mansoor, S.; Bedford, I.D.; Pinner, M.S.; Saunders, K.; Stanley, J.; Zafar, Y.; Malik, K.A.; Markham, P.G. Identification of DNA components required for induction of cotton leaf curl disease. Virology 2001, 285, 234–243. [Google Scholar] [CrossRef] [PubMed]
- Rojas, M.R.; Hagen, C.; Lucas, W.J.; Gilbertson, R.L. Exploiting chinks in the plant’s armor: Evolution and emergence of geminiviruses. Annu. Rev. Phytopathol. 2005, 43, 361–394. [Google Scholar] [CrossRef] [PubMed]
- Martin, D.P.; Lefeuvre, P.; Varsani, A.; Hoareau, M.; Semegni, J.Y.; Dijoux, B.; Vincent, C.; Lett, J.M. Complex recombination patterns arising during geminivirus co infections both preserve and demarcate biologically important intra-genome interaction networks. PLoS Pathog. 2011, 7, e1002203. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saunders, K.; Bedford, I.D.; Stanley, J. Pathogenicity of a natural recombinant associated with Ageratum yellow vein disease: Implications for geminivirus evolution and disease aetiology. Virology 2001, 282, 38–47. [Google Scholar] [CrossRef] [PubMed]
- Tao, X.R.; Zhou, X.P. Pathogenicity of a naturally occurring recombinant DNA satellite associated with tomato yellow leaf curl china virus. J. Gen. Virol. 2008, 89, 306–311. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Xie, Y.; Zhang, L.; Ren, H.; Li, Z. A naturally occurring defective DNA satellite associated with a monopartite begomovirus: Evidence for recombination between alphasatellite and betasatellite. Viruses 2013, 5, 2116–2128. [Google Scholar] [CrossRef] [PubMed]
- Pilartz, M.; Jeske, H. Abutilon mosaic geminivirus double-stranded DNA is packed into minichromosomes. Virology 1992, 189, 800–802. [Google Scholar] [CrossRef]
- Gutierrez, C. Geminivirus DNA replication. Cell. Mol. Life Sci. 1999, 56, 313–329. [Google Scholar] [CrossRef] [PubMed]
- Briddon, R.W.; Markham, P.G. Cotton leaf curl virus disease. Virus Res. 2001, 71, 151–159. [Google Scholar] [CrossRef]
- Thresh, J.M.; Cooter, R.J. Strategies for controlling cassava mosaic virus disease in Africa. Plant Pathol. 2005, 54, 587–614. [Google Scholar] [CrossRef]
- Shepherd, D.N.; Martin, D.P.; Van der Walt, E.; Dent, K.; Varsani, A.; Rybicki, E.P. Maize streak virus: An old and complex ‘emerging’ pathogen. Mol. Plant Pathol. 2010, 11, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Varma, A.; Malathi, V.G. Emerging geminivirus problems: A serious threat to crop production. Ann. Appl. Biol. 2003, 142, 145–164. [Google Scholar] [CrossRef]
- Moffat, A.S. Geminiviruses emerge as serious crop threat. Science 1999, 286, 1835. [Google Scholar] [CrossRef]
- Lefeuvre, P.; Martin, D.P.; Harkins, G.; Lemey, P.; Gray, A.J.A.; Meredith, S.; Lakay, F.; Monjane, A.; Lett, J.-M.; Varshini, A.; et al. The Spread of Tomato Yellow Leaf Curl Virus from the Middle East to the World. PLoS Pathog. 2010, 6, e1001164. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shivaprasad, P.V.; Akbergenov, R.; Trinks, D.; Rajeswaran, R.; Veluthambi, K.; Hohn, T.; Pooggin, M.M. Promoters, transcripts, and regulatory proteins of mungbean yellow mosaic geminivirus. J. Virol. 2005, 79, 8149–8163. [Google Scholar] [CrossRef] [PubMed]
- Akbergenov, R.; Si-Ammour, A.; Blevins, T.; Amin, I.; Kutter, C.; Vanderschuren, H.; Zhang, P.; Gruissem, W.; Meins, F., Jr.; Hohn, T.; et al. Molecular characterization of geminivirus-derived small RNAs in different plant species. Nucleic Acids Res. 2006, 34, 462–471. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Negrete, E.A.; Carrillo-Tripp, J.; Rivera-Bustamante, R.F. RNA silencing against geminivirus: Complementary action of posttranscriptional gene silencing and transcriptional gene silencing in host recovery. J. Virol. 2009, 83, 1332–1340. [Google Scholar] [CrossRef] [PubMed]
- Muthamilarasan, M.; Prasad, M. Plant innate immunity: An updated insight into defense mechanism. J. Biosci. 2013, 38, 433–449. [Google Scholar] [CrossRef] [PubMed]
- Okamura, K.; Ishizuka, A.; Siomi, H.; Siomi, M.C. Distinct roles for Argonaute proteins in small RNA-directed RNA cleavage pathways. Genes Dev. 2004, 18, 1655–1666. [Google Scholar] [CrossRef] [PubMed]
- Yu, B.; Yang, Z.; Li, J.; Minakhina, S.; Yang, M.; Padgett, R.W.; Steward, R.; Chen, X. Methylation as a crucial step in plant microRNA biogenesis. Science 2005, 307, 932–935. [Google Scholar] [CrossRef] [PubMed]
- Brough, C.L.; Gardiner, W.E.; Inamdar, N.M.; Zhang, X.Y.; Ehrlich, M.; Bisaro, D.M. DNA methylation inhibits propagation of tomato golden mosaic virus DNA in transfected protoplasts. Plant Mol. Biol. 1992, 18, 703–712. [Google Scholar] [CrossRef] [PubMed]
- Seemanpillai, M.; Dry, I.; Randles, J.; Rezaian, A. Transcriptional silencing of geminiviral promoter-driven transgenes following homologous virus infection. Mol. Plant-Microbe Interact. 2003, 16, 429–438. [Google Scholar] [CrossRef] [PubMed]
- Pooggin, M.; Hohn, T. RNAi targeting of DNA virus in plants. Nat. Biotechnol. 2003, 21, 131–132. [Google Scholar] [CrossRef] [PubMed]
- Raja, P.; Sanville, B.C.; Buchmann, R.C.; Bisaro, D.M. Viral genome methylation as an epigenetic defense against geminiviruses. J. Virol. 2008, 82, 8997. [Google Scholar] [CrossRef] [PubMed]
- Sunter, G.; Sunter, J.; Bisaro, D.M. Plants expressing tomato golden mosaic virus AL2 or beet curly top virus L2 transgenes show enhanced susceptibility to infection by DNA and RNA viruses. Virology 2001, 285, 59–70. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Hao, L.; Shung, C.Y.; Sunter, G.; Bisaro, D.M. Adenosine kinase is inactivated by geminivirus AL2 and L2 proteins. Plant Cell 2003, 15, 3020–3032. [Google Scholar] [CrossRef] [PubMed]
- Hagen, C.; Rojas, M.R.; Kon, T.; Gilbertson, R.L. Recovery from cucurbit leaf crumple virus (family Geminiviridae, genus Begomovirus) infection is an adaptive antiviral response associated with changes in viral small RNAs. Phytopathology 2008, 98, 1029–1037. [Google Scholar] [CrossRef] [PubMed]
- Carrillo-Tripp, J.; Lozoya-Gloria, E.; Rivera-Bustamante, R.F. Symptom remission and specific resistance of pepper plants after infection by pepper golden mosaic virus. Phytopathology 2007, 97, 51–57. [Google Scholar] [CrossRef] [PubMed]
- Havecker, E.R.; Wallbridge, L.M.; Hardcastle, T.J.; Bush, M.S.; Kelly, K.A.; Dunn, R.M.; Schwach, F.; Doonan, J.H.; Baulcombe, D.C. The Arabidopsis RNA-directed DNA methylation argonautes functionally diverge based on their expression and interaction with target loci. Plant Cell 2010, 22, 321–334. [Google Scholar] [CrossRef] [PubMed]
- Ding, S.W.; Voinnet, O. Antiviral immunity directed by small RNAs. Cell 2007, 130, 413–426. [Google Scholar] [CrossRef] [PubMed]
- Carrington, J.C.; Ambros, V. Role of microRNAs in plant and animal development. Science 2003, 301, 336–338. [Google Scholar] [CrossRef] [PubMed]
- Baulcombe, D.C. RNA silencing in plants. Nature 2004, 431, 356–363. [Google Scholar] [CrossRef] [PubMed]
- Matzke, M.A.; Birchler, J.A. RNAi-mediated pathways in the nucleus. Nat. Rev. Genet. 2005, 6, 24–35. [Google Scholar] [CrossRef] [PubMed]
- Vazquez, F.; Legrand, S.; Windels, D. The biosynthetic pathways and biological scopes of plant small RNAs. Trends Plant Sci. 2010, 15, 337–345. [Google Scholar] [CrossRef] [PubMed]
- Blevins, T.; Rajeswaran, R.; Shivaprasad, P.V.; Beknazariants, D.; Si-Ammour, A.; Park, H.S.; Vazquez, F.; Robertson, D.; Meins, F., Jr.; Hohn, T.; et al. Four plant Dicers mediate viral small RNA biogenesis and DNA virus induced silencing. Nucleic Acids Res. 2006, 34, 6233–6246. [Google Scholar] [CrossRef] [PubMed]
- Vanderschuren, H.; Akbergenov, R.; Pooggin, M.M.; Hohn, T.; Gruissem, W.; Zhang, P. Transgenic cassava resistance to African cassava mosaic virus is enhanced by viral DNAA bidirectional promoter-derived siRNAs. Plant Mol. Biol. 2007, 64, 549–557. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Wang, Y.; Guo, W.; Xie, Y.; Xie, Q.; Fan, L.; Zhou, X. Characterization of small interfering RNAs derived from the geminivirus/β satellite complex using deep sequencing. PLoS ONE 2011, 6, e16928. [Google Scholar]
- Sahu, P.P.; Sharma, N.; Puranik, S.; Prasad, M. Post-transcriptional and epigenetic arms of RNA silencing: A defense machinery of naturally tolerant tomato plant against Tomato leaf curl New Delhi virus. Plant Mol. Biol. Rep. 2014, 32, 1015–1029. [Google Scholar] [CrossRef]
- Paprotka, T.; Deuschle, K.; Metzler, V.; Jeske, H. Conformation-selective methylation of Geminivirus DNA. J. Virol. 2011, 85, 12001–12012. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Ruiz, H.; Takeda, A.; Chapman, E.J.; Sullivan, C.M.; Fahlgren, N.; Brempelis, K.J.; Carrington, J.C. Arabidopsis RNA-dependent RNA polymerases and dicer-like proteins in antiviral defense and small interfering RNA biogenesis during Turnip mosaic virus infection. Plant Cell 2010, 22, 481–496. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.B.; Jovel, J.; Udomporn, P.; Wang, Y.; Wu, Q.F.; Li, W.X.; Gasciolli, V.; Vaucheret, H.; Ding, S.W. The 21-nucleotide, but not 22-nucleotide, viral secondary small interfering RNAs direct potent antiviral defense by two cooperative Argonautes in Arabidopsis thaliana. Plant Cell 2011, 23, 1625–1638. [Google Scholar] [CrossRef] [PubMed]
- Aregger, M.; Borah, B.K.; Seguin, J.; Rajeswaran, R.; Gubaeva, E.G.; Zvereva, A.S.; Windels, D.; Vazquez, F.; Blevins, T.; Farinelli, L.; et al. Primary and secondary siRNAs in geminivirus-induced gene silencing. PLoS Pathog. 2012, 8, e1002941. [Google Scholar] [CrossRef] [PubMed]
- Hanley-Bowdoin, L.; Bejarano, E.R.; Robertson, D.; Mansoor, S. Geminiviruses: Masters at redirecting and reprogramming plant processes. Nat. Rev. Microbiol. 2013, 11, 777–788. [Google Scholar] [CrossRef] [PubMed]
- Raja, P.; Jackel, J.N.; Li, S.; Heard, I.M.; Bisaro, D.M. Arabidopsis Double-stranded RNA binding protein DRB3 participates in methylation-mediated defense against Geminiviruses. J. Virol. 2014, 88, 2611–2622. [Google Scholar] [CrossRef] [PubMed]
- Naqvi, A.R.; Haq, Q.M.; Mukherjee, S.K. MicroRNA profiling of Tomato leaf curl New Delhi virus (ToLCNDV) infected tomato leaves indicates that deregulation of mir159/319 and mir172 might be linked with leaf curl disease. Virol. J. 2010, 7, 281. [Google Scholar] [CrossRef] [PubMed]
- Amin, I.; Basavaprabhu, L.P.; Briddon, R.W.; Mansoor, S.; Fauquet, C.M. Common set of developmental miRNAs are upregulated in Nicotiana benthamiana by diverse begomoviruses. Virol. J. 2011, 8, 143. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Quintero, A.; Neme, R.; Zapata, A.; López, C. Plant miRNAs and their role in defense against viruses: A bioinformatics approach. BMC Plant Biol. 2010, 10, 138. [Google Scholar] [CrossRef] [PubMed]
- Ramesh, S.V.; Chouhan, B.S.; Gaurav, K.; Praveen, S.; Chand, S. Expression dynamics of Glycine max (L.) Merrill derived microRNAs (miRNAs) and their targets during Mungbean yellow mosaic India virus (MYMIV) infection. Physiol. Mol. Plant Pathol. 2017, 100, 13–22. [Google Scholar] [CrossRef]
- Tousi, N.; Eini, O.; Ahmadvand, R.; Carra, A.; Miozzi, L.; Noris, E.; Accotto, G.P. In silico prediction of miRNAs targeting ToLCV and their regulation in susceptible and resistant tomato plants. Aust. Plant Pathol. 2017, 46, 379–386. [Google Scholar] [CrossRef]
- Ramesh, S.V.; Ratnaparkhe, M.B.; Kumawat, G.; Gupta, G.K.; Husain, S.M. Plant miRNAome and antiviral resistance: A retrospective view and prospective challenges. Virus Genes 2014, 48, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Chellappan, P.; Vanitharani, R.; Ogbe, F.; Fauquet, C.M. Effect of temperature on geminivirus-induced RNA silencing in plants. Plant Physiol. 2005, 138, 1828–1841. [Google Scholar] [CrossRef] [PubMed]
- Maghuly, F.; Ramkat, R.C.; Laimer, M. Virus versus host plant microRNAs: Who determines the outcome of the interaction? PLoS ONE 2014, 9, e98263. [Google Scholar] [CrossRef] [PubMed]
- Voinnet, O.; Pinto, Y.M.; Baulcombe, D.C. Suppression of gene silencing: A general strategy used by diverse DNA and RNA viruses of plants. Proc. Natl. Acad. Sci. USA 1999, 96, 14147–14152. [Google Scholar] [CrossRef] [PubMed]
- Van Wezel, R.; Liu, H.; Tien, P.; Stanley, J.; Hong, Y. Mutation of three cysteine residues in Tomato yellow leaf curl virus-China C2 protein causes dysfunction in pathogenesis and posttranscriptional gene-silencingsuppression. Mol. Plant-Microbe Interact. 2002, 15, 203–208. [Google Scholar] [CrossRef] [PubMed]
- Dong, X.; van Wezel, R.; Stanley, J.; Hong, Y. Functional characterization of the nuclear localization signal for a suppressor of posttranscriptional gene silencing. J. Virol. 2003, 77, 7026–7033. [Google Scholar] [CrossRef] [PubMed]
- Trinks, D.; Rajeswaran, R.; Shivaprasad, P.V.; Akbergenov, R.; Oakeley, E.J.; Veluthambi, K.; Hohn, T.; Pooggin, M. Suppression of RNA silencing by a geminivirus nuclear protein, AC2, correlates with transactivation of host genes. J. Virol. 2005, 79, 2517–2527. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Buckley, K.J.; Yang, X.; Buchmann, R.C.; Bisaro, D.M. Adenosine kinase inhibition and suppression of RNA silencing by geminivirus AL2 and L2 proteins. J. Virol. 2005, 79, 7410–7418. [Google Scholar] [CrossRef] [PubMed]
- Buchmann, R.C.; Asad, S.; Wolf, J.N.; Mohannath, G.; Bisaro, D.M. Geminivirus AL2 and L2 proteins suppress transcriptional gene silencing and cause genome-wide reductions in cytosine methylation. J. Virol. 2009, 83, 5005–5013. [Google Scholar] [CrossRef] [PubMed]
- Baliji, S.; Sunter, J.; Sunter, G. Transcriptional analysis of complementary sense genes in Spinach curly top virus and functional role of C2 in pathogenesis. Mol. Plant-Microbe Interact. 2007, 20, 194–206. [Google Scholar] [CrossRef] [PubMed]
- Jackel, J.N.; Buchmann, R.C.; Singhal, U.; Bisaro, D.M. Analysis of Geminivirus AL2 and L2 Proteins Reveals a Novel AL2 Silencing Suppressor Activity. J. Virol. 2015, 89, 3176–3187. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Chen, H.; Huang, X.; Xia, R.; Zhao, Q.; Lai, J.; Teng, K.; Li, Y.; Liang, L.; Du, Q.; et al. BSCTV C2 attenuates the degradation of SAMDC 1 to suppress DNA methylation-mediated gene silencing in Arabidopsis. Plant Cell 2011, 23, 273–288. [Google Scholar] [CrossRef] [PubMed]
- Vanitharani, R.; Chellappan, P.; Pita, J.S.; Fauquet, C. Differential roles of AC2 and AC4 of cassava geminiviruses in mediating synergism and suppression of posttranscriptional gene silencing. J. Virol. 2004, 78, 9487–9498. [Google Scholar] [CrossRef] [PubMed]
- Vanitharani, R.; Chellappan, P.; Fauquet, C.M. Geminiviruses and RNA silencing. Trends Plant Sci. 2005, 10, 144–151. [Google Scholar] [CrossRef] [PubMed]
- Bazzini, A.A.; Hopp, H.E.; Beachy, R.N.; Asurmendi, S. Infection and co-accumulation of tobacco mosaic virus proteins alter microRNA levels, correlating with symptom and plant development. Proc. Natl. Acad. Sci. USA 2007, 104, 12157–12162. [Google Scholar] [CrossRef] [PubMed]
- Amin, I.; Hussain, K.; Akbergenov, R.; Yadav, J.S.; Qazi, J.; Mansoor, S.; Hohn, T.; Fauquet, C.M.; Briddon, R.W. Suppressors of RNA silencing encoded by the components of the cotton leaf curl begomovirus-beta-satellite complex. Mol. Plant-Microbe Interact. 2011, 24, 973–983. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Xu, X.; Huang, C.; Gu, Z.; Cao, L.; Hu, T.; Ding, M.; Li, Z.; Zhou, X. The AC5 protein encoded by Mungbean yellow mosaic India virus is a pathogenicity determinant that suppresses RNA silencing-based antiviral defenses. New Phytol. 2015, 208, 555–569. [Google Scholar] [CrossRef] [PubMed]
- Zrachya, A.; Glick, E.; Levy, Y.; Arazi, T.; Citovsky, V.; Gafni, Y. Suppressor of RNA silencing encoded by Tomato yellow leaf curl virus-Israel. Virology 2007, 358, 159–165. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Dong, J.; Xu, Y.; Wu, J. V2 protein encoded by Tomato yellow leaf curl China virus is an RNA silencing suppressor. Virus Res. 2012, 163, 51–58. [Google Scholar] [CrossRef] [PubMed]
- Glick, E.; Zrachya, A.; Levy, Y.; Mett, A.; Gidoni, D.; Belausov, E.; Citovsky, V.; Gafni, Y. Interaction with host SGS3 is required for suppression of RNA silencing by tomato yellow leaf curl virus V2 protein. Proc. Natl. Acad. Sci. USA 2008, 105, 157–161. [Google Scholar] [CrossRef] [PubMed]
- Cui, X.; Li, G.; Wang, D.; Hu, D.; Zhou, X. A begomovirus DNAβ-encoded protein binds DNA, functions as a suppressor of RNA silencing, and targets the cell nucleus. J. Virol. 2005, 79, 10764–10775. [Google Scholar] [CrossRef] [PubMed]
- Bisaro, D.M. Silencing suppression by geminivirus proteins. Virology 2006, 344, 158–168. [Google Scholar] [CrossRef] [PubMed]
- Gopal, P.; Kumar, P.P.; Sinilal, B.; Jose, J.; Yadunandam, A.K.; Usha, R. Differential roles of C4 and βC1 in mediating suppression of post-transcriptional gene silencing: Evidence for transactivation by the C2 of Bhendi yellow vein mosaic virus, a monopartite begomovirus. Virus Res. 2007, 123, 9–18. [Google Scholar] [CrossRef] [PubMed]
- Kon, T.; Sharma, P.; Ikegami, M. Suppressor of RNA silencing encoded by the monopartite tomato leaf curl Java Begomovirus. Arch. Virol. 2007, 152, 1273–1282. [Google Scholar] [CrossRef] [PubMed]
- Shukla, R.; Dalal, S.; Malathi, V.G. Suppressors of RNA silencing encoded by tomato leaf curl betasatellites. J. Biosci. 2013, 38, 1–7. [Google Scholar] [CrossRef]
- Li, F.; Huang, C.; Li, Z.; Zhou, X. Suppression of RNA Silencing by a Plant DNA Virus Satellite Requires a Host Calmodulin-Like Protein to Repress RDR6 Expression. PLoS Pathog. 2014, 10, e1003921. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Dang, M.; Hou, H.; Mei, Y.; Qian, Y.; Zhou, X. Identification of an RNA silencing suppressor encoded by a Mastrevirus. J. Gen. Virol. 2014, 95, 2082–2088. [Google Scholar] [CrossRef] [PubMed]
- Nawaz-ul-Rehman, M.S.; Nahid, N.; Mansoor, S.; Briddon, R.W.; Fauquet, C.M. Post-transcriptional gene silencing suppressor activity of two non-pathogenic alphasatellites associated with a begomovirus. Virology 2010, 405, 300–308. [Google Scholar] [CrossRef] [PubMed]
- Hanley-Bowdoin, L.; Settlage, S.B.; Robertson, D. Reprogramming plant gene expression: A prerequisite to geminivirus DNA replication. Mol. Plant Pathol. 2004, 5, 149–156. [Google Scholar] [CrossRef] [PubMed]
- Sahu, P.P.; Sharma, N.; Puranik, S.; Muthamilarasan, M.; Prasad, M. Involvement of host regulatory pathways during geminivirus infection: A novel platform for generating durable resistance. Funct. Integr. Genom. 2014, 14, 47–58. [Google Scholar] [CrossRef] [PubMed]
- Castillo, A.G.; Collinet, D.; Deret, S.; Kashoggi, A.; Bejarano, E.R. Dual interaction of plant PCNA with geminivirus replication accessory protein (REn) and viral replication protein (Rep). Virology 2003, 312, 381–394. [Google Scholar] [CrossRef]
- Bagewadi, B.; Chen, S.; Lal, S.K.; Choudhury, N.R.; Mukherjee, S.K. PCNA interacts with Indian mung bean yellow mosaic virus Rep and downregulates Rep activity. J. Virol. 2004, 78, 11890–11903. [Google Scholar] [CrossRef] [PubMed]
- Settlage, S.B.; See, R.G.; Hanley-Bowdoin, L. Geminivirus C3 protein: Replication enhancement and protein interactions. J. Virol. 2005, 79, 9885–9895. [Google Scholar] [CrossRef] [PubMed]
- Bruce, G.; Gu, M.; Shi, N.; Liu, Y.; Hong, Y. Influence of retinoblastoma-related gene silencing on the initiation of DNA replication by African cassava mosaic virus Rep in cells of mature leaves in Nicotiana benthamiana plants. Virol. J. 2011, 8, 561. [Google Scholar] [CrossRef] [PubMed]
- Singh, D.K.; Islam, M.N.; Choudhury, N.R.; Karjee, S.; Mukherjee, S.K. The 32 kDa subunit of replication protein A (RPA) participates in the DNA replication of Mungbean yellow mosaic India virus (MYMIV) by interacting with the viral Rep protein. Nucleic Acids Res. 2007, 35, 755–770. [Google Scholar] [CrossRef] [PubMed]
- Lozano-Durán, R.; Rosas-Díaz, T.; Luna, A.P.; Bejarano, E.R. Identification of host genes involved in geminivirus infection using a reverse genetics approach. PLoS ONE 2011, 6, e22383. [Google Scholar] [CrossRef] [PubMed]
- Suyal, G.; Rana, V.P.; Mukherjee, S.K.; Saima, W.; Choudhury, N.R. Arabidopsis thaliana NAC083 protein interacts with Mungbean yellow mosaic India virus (MYMIV) Rep protein. Virus Genes 2014, 48, 486–493. [Google Scholar] [CrossRef] [PubMed]
- Ascencio-Ibáñez, J.T.; Sozzani, R.; Lee, T.J.; Chu, T.M.; Wolfinger, R.D.; Cella, R.; Hanley-Bowdoin, L. Global analysis of Arabidopsis gene expression uncovers a complex array of changes impacting pathogen response and cell cycle during geminivirus infection. Plant Physiol. 2008, 148, 436–454. [Google Scholar] [CrossRef] [PubMed]
- Hao, L.; Wang, H.; Sunter, G.; Bisaro, D.M. Geminivirus AL2 and L2 proteins interact with and inactivate SNF1 kinase. Plant Cell 2003, 15, 1034–1048. [Google Scholar] [CrossRef] [PubMed]
- Florentino, L.H.; Santos, A.A.; Fontenelle, M.R.; Pinheiro, G.L.; Zerbini, F.M.; Baracat-Pereira, M.C.; Fontes, E.P. A PERK-like receptor kinase interacts with the geminivirus nuclear shuttle protein and potentiates viral infection. J. Virol. 2006, 80, 6648–6656. [Google Scholar] [CrossRef] [PubMed]
- Fregene, M.; Matsumura, H.; Akano, A.; Dixon, A.; Terauchi, R. Serial analysis of gene expression (SAGE) of host-plant resistance to the cassava mosaic disease (CMD). Plant Mol. Biol. 2004, 56, 563–571. [Google Scholar] [CrossRef] [PubMed]
- Egelkrout, E.M.; Robertson, D.; Hanley-Bowdoin, L. Proliferating cell nuclear antigen transcription is repressed through an E2F consensus element and activated by geminivirus infection in mature leaves. Plant Cell 2001, 13, 1437–1452. [Google Scholar] [CrossRef] [PubMed]
- Marathe, R.; Guan, Z.; Anandalakshmi, R.; Zhao, H.; Dinesh-Kumar, S.P. Study of Arabidopsis thaliana resistome in response to cucumber mosaic virus infection using whole genome microarray. Plant Mol. Biol. 2004, 55, 501–520. [Google Scholar] [CrossRef] [PubMed]
- Kaur, H.; Yadav, C.; Alatar, A.; Faisal, M.; Jyothsna, P.; Malathi, V.G.; Praveen, S. Gene expression changes in tomato during symptom development in response to leaf curl virus infection. J. Plant Biochem. Biotechnol. 2014, 24, 347–354. [Google Scholar] [CrossRef]
- Mockaitis, K.; Estelle, M. Auxin receptors and plant development: A new signaling paradigm. Annu. Rev. Cell Dev. Biol. 2008, 24, 55–80. [Google Scholar] [CrossRef] [PubMed]
- Schommer, C.; Palatnik, J.F.; Aggarwal, P.; Chetelat, A.; Cubas, P.; Farmer, E.E.; Nath, U.; Weigel, D. Control of jasmonate biosynthesis and senescence by miR319 targets. PLoS Biol. 2008, 6, 1991–2001. [Google Scholar] [CrossRef] [PubMed]
- Rubio-Somoza, I.; Cuperus, J.T.; Weigel, D.; Carrington, J.C. Regulation and functional specialization of small RNA-target nodes during plant development. Curr. Opin. Plant Biol. 2009, 12, 622–627. [Google Scholar] [CrossRef] [PubMed]
- Fondong, V.F.; Felton, C.; Reddy, R.V.C.; Lu, C.; Hankoua, B.; Czymmek, K.; Achenjang, F. The consensus N-myristoylation motif of a geminivirus AC4 protein is required for membrane binding and pathogenicity. Mol. Plant-Microbe Interact. 2007, 20, 380–391. [Google Scholar] [CrossRef] [PubMed]
- Fontes, E.P.; Santos, A.A.; Luz, D.F.; Waclawovsky, A.J.; Chory, J. The geminivirus nuclear shuttle protein is a virulence factor that suppresses transmembrane receptor kinase activity. Genes Dev. 2004, 18, 2545–2556. [Google Scholar] [CrossRef] [PubMed]
- Piroux, N.; Saunders, K.; Page, A.; Stanley, J. Geminivirus pathogenicity protein C4 interacts with Arabidopsis thaliana shaggy-related protein kinase AtSKeta, a component of the brassinosteroid signaling pathway. Virology 2007, 362, 428–440. [Google Scholar] [CrossRef] [PubMed]
- Lewis, J.D.; Lazarowitz, S.G. Arabidopsis synaptotagmin SYTA regulates endocytosis and virus movement protein cell-to-cell transport. Proc. Natl. Acad. Sci. USA 2010, 107, 2491–2496. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, C.M.; Fontenelle, M.R.; Florentino, L.H.; Santos, A.A.; Zerbini, F.M.; Fontes, E.P. A novel nucleocytoplasmic traffic GTPase identified as a functional target of the bipartite geminivirus nuclear shuttle protein. Plant J. 2008, 55, 869–880. [Google Scholar] [CrossRef] [PubMed]
- Eini, O.; Dogra, S.; Selth, L.A.; Dry, I.B.; Randles, J.W.; Rezaian, M.A. Interaction with a host ubiquitin-conjugating enzyme is required for the pathogenicity of a geminiviral DNA beta satellite. Mol. Plant-Microbe Interact. 2009, 22, 737–746. [Google Scholar] [CrossRef] [PubMed]
- Castillo, A.G.; Kong, L.J.; Hanley-Bowdoin, L.; Bejarano, E.R. Interaction between a geminivirus replication protein and the plant sumoylation system. J. Virol. 2004, 78, 2758–2769. [Google Scholar] [CrossRef] [PubMed]
- Czosnek, H.; Eybishtz, A.; Sade, D.; Gorovits, R.; Sobol, I.; Bejarano, E.; Rosas-Díaz, T.; Lozano-Durán, R. Discovering host genes involved in the infection by the tomato yellow leaf curl virus complex and in the establishment of resistance to the virus using tobacco rattle virus-based post transcriptional gene silencing. Viruses 2013, 5, 998–1022. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, F.; Zhao, N.; Li, Z.; Xu, X.; Wang, Y.; Yang, X.; Liu, S.-S.; Wang, A.; Zhou, X. A calmodulin-like protein suppresses RNA silencing and promotes geminivirus infection by degrading SGS3 via the autophagy pathway in Nicotiana benthamiana. PLoS Pathog. 2017, 13, e1006213. [Google Scholar] [CrossRef] [PubMed]
- Schoelz, J.E.; Harries, P.A.; Nelson, R.S. Intracellular transport of plant viruses: Finding the door out of the cell. Mol. Plant 2011, 4, 813–831. [Google Scholar] [CrossRef] [PubMed]
- Bar-Ziv, A.; Levy, Y.; Hak, H.; Mett, A.; Belausov, E.; Citovsky, V.; Gafni, Y. The Tomato yellow leaf curl virus (TYLCV) V2 protein interacts with the host papain-like cysteine protease CYP1. Plant Signal Behav. 2012, 7, 83–989. [Google Scholar] [CrossRef] [PubMed]
- Miozzi, L.; Napoli, C.; Sardo, L.; Accotto, G.P. Transcriptomics of the interaction between the monopartite phloem-limited geminivirus Tomato Yellow Leaf Curl Sardinia Virus and Solanum lycopersicum highlights a role for plant hormones, autophagy and plant immune system fine tuning during Infection. PLoS ONE 2014, 9, e89951. [Google Scholar] [CrossRef] [PubMed]
- Mills-Lujan, K.; Andrews, D.L.; Chou, C.W.; Deom, C.M. The roles of phosphorylation and SHAGGY-like Protein Kinases in geminivirus C4 protein induced hyperplasia. PLoS ONE 2015, 10, e0122356. [Google Scholar] [CrossRef] [PubMed]
- Ramesh, S.V.; Mishra, A.K.; Praveen, S. Hairpin RNA-mediated strategies for silencing of Tomato leaf curl virus AC1 and AC4 genes for effective resistance in plants. Oligonucleotides 2007, 17, 251–257. [Google Scholar] [CrossRef] [PubMed]
- Praveen, S.; Ramesh, S.V.; Mishra, A.K.; Koundal, V.; Palukaitis, P. Silencing potential of viral derived RNAi constructs in Tomato leaf curl virus-AC4 gene suppression in tomato. Transgenic Res. 2010, 19, 45–55. [Google Scholar] [CrossRef] [PubMed]
- Niu, Q.W.; Lin, S.S.; Reyes, J.L.; Chen, K.C.; Wu, H.W.; Yeh, S.D.; Chua, N.H. Expression of artificial microRNAs in transgenic Arabidopsis thaliana confers virus resistance. Nat. Biotechnol. 2006, 24, 1420–1428. [Google Scholar] [CrossRef] [PubMed]
- Qu, J.; Ye, J.; Fang, R. Artificial microRNA-mediated virus resistance in plants. J. Virol. 2007, 81, 6690–6699. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Taneja, J.; Dasgupta, I.; Mukherjee, S.K. Development of plants resistant to tomato geminiviruses using artificial trans-acting small interfering RNA. Mol. Plant Pathol. 2015, 16, 724–734. [Google Scholar] [CrossRef] [PubMed]
- Verlaan, M.G.; Hutton, S.F.; Ibrahem, R.M.; Kormelink, R.; Visser, R.G.F.; Scott, J.W.; Bai, Y.L. The Tomato Yellow Leaf Curl Virus Resistance Genes Ty-1 and Ty-3 Are Allelic and Code for DFDGD-Class RNA-Dependent RNA Polymerases. PLoS Genet. 2013, 9, e1003399. [Google Scholar] [CrossRef] [PubMed]
- Vanderschuren, H.; Moreno, I.; Anjanappa, R.B.; Zainuddin, I.M.; Gruissem, W. Exploiting the Combination of Natural and Genetically Engineered Resistance to Cassava Mosaic and Cassava Brown Streak Viruses Impacting Cassava Production in Africa. PLoS ONE 2012, 7, e45277. [Google Scholar] [CrossRef] [PubMed]
- Reyes, M.I.; Nash, T.E.; Dallas, M.M.; Ascencio-Ibáñez, J.T.; Hanley-Bowdoin, L. Peptide aptamers that bind to geminivirus replication proteins confer a resistance phenotype to Tomato yellow leaf curl virus and Tomato mottle virus infection in tomato. J. Virol. 2013, 87, 9691–9706. [Google Scholar] [CrossRef] [PubMed]
- Sansregret, R.; Dufour, V.; Langlois, M.; Daayf, F.; Dunoyer, P.; Voinnet, O.; Bouarab, K. Extreme Resistance as a Host Counter-counter Defense against Viral Suppression of RNA Silencing. PLoS Pathog. 2013, 9, e1003435. [Google Scholar] [CrossRef] [PubMed]
- Baltes, N.J.; Hummel, A.W.; Konecna, E.; Cegan, R.; Bruns, A.N.; Bisaro, D.M.; Voytas, D.F. Conferring resistance to geminiviruses with the CRISPR–Cas prokaryotic immune system. Nat. Plants 2015, 1, 15145. [Google Scholar] [CrossRef]
- Ji, X.; Zhang, H.; Zhang, Y.; Wang, Y.; Gao, C. Establishing a CRISPR–Cas-like immune system conferring DNA virus resistance in plants. Nat.Plants 2015, 1, 15144. [Google Scholar] [CrossRef] [PubMed]
- Ali, Z.; Abulfaraj, A.; Idris, A.; Ali, S.; Tashkandi, M.; Mahfouz, M.M. CRISPR/Cas9-mediated viral interference in plants. Genome Biol. 2015, 16, 238. [Google Scholar] [CrossRef] [PubMed]
- Ali, Z.; Ali, S.; Tashkandi, M.; Zaidi, S.S.E.A.; Mahfouz, M.M. CRISPR/Cas9-mediated immunity to geminiviruses: Differential interference and evasion. Sci.Rep. 2016, 6, 26912. [Google Scholar] [CrossRef] [PubMed]
- Sahu, P.P.; Rai, N.K.; Chakraborty, S.; Singh, M.; Chandrappa, P.H.; Ramesh, B.; Chattopadhyay, D.; Prasad, M. Tomato cultivar tolerant to Tomato leaf curl New Delhi virus infection induces virus-specific short interfering RNA accumulation and defense-associated host gene expression. Mol. Plant Pathol. 2010, 11, 531–544. [Google Scholar] [CrossRef] [PubMed]
- Yadav, R.K.; Chattopadhyay, D. Enhanced viral intergenic region–specific short interfering RNA accumulation and DNA methylation correlates with resistance against a Geminivirus. Mol. Plant-Microbe Interact. 2011, 24, 1189–1197. [Google Scholar] [CrossRef] [PubMed]
- Sahu, P.P.; Rai, N.K.; Puranik, S.; Roy, A.; Khan, M.; Prasad, M. Dynamics of defense-related components in two contrasting genotypes of tomato upon infection with Tomato leaf curl New Delhi virus. Mol. Biotechnol. 2012, 52, 140–150. [Google Scholar] [CrossRef] [PubMed]
- Pandey, P.; Choudhury, N.R.; Mukherjee, S.K. A geminiviral amplicon (VA) derived from Tomato leaf curl virus (ToLCV) can replicate in a wide variety of plant species and also acts as a VIGS vector. Virol. J. 2009, 6, 152. [Google Scholar] [CrossRef] [PubMed]
- Mor, T.S.; Moon, Y.S.; Palmer, K.E.; Mason, H.S. Geminivirus vectors for high-level expression of foreign proteins in plant cells. Biotechnol. Bioeng. 2003, 81, 430–437. [Google Scholar] [CrossRef] [PubMed]
- Regnard, G.L.; Halley-Stott, R.P.; Tanzer, F.L.; Hitzeroth, I.I.; Rybicki, E.P. High level protein expression in plants through the use of a novel autonomously replicating geminivirus shuttle vector. Plant Biotechnol. J. 2010, 8, 38–46. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.G.; Cha, J.; Chandrasegaran, S. Hybrid restriction enzymes: Zinc finger fusions to Fok Icleavage domain. Proc. Natl. Acad. Sci. USA 1996, 93, 1156–1160. [Google Scholar] [CrossRef] [PubMed]
- Miller, J.C.; Siyuan, T.; Guijuan, Q.; Kyle, A.B.; Wang, J.; Xia, D.F.; Meng, X.; Paschon, D.E.; Leung, E.; Hinkley, S.J.; et al. A TALE nuclease architecture for efficient genome editing. Nat. Biotechnol. 2011, 29, 143–148. [Google Scholar] [CrossRef] [PubMed]
- Baltes, N.J.; Gil-Humanes, J.; Cermak, T.; Atkins, P.A.; Voytas, D.F. DNA replicons for plant genome engineering. Plant Cell 2014, 26, 151–163. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Lu, Y.; Botella, J.; Mao, Y.; Hua, K.; Zhu, J.K. Gene Targeting by Homology-directed Repair in Rice using a Geminivirus-based CRISPR/Cas9 System. Mol. Plant 2017, 10, 1007–1010. [Google Scholar] [CrossRef] [PubMed]

© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ramesh, S.V.; Sahu, P.P.; Prasad, M.; Praveen, S.; Pappu, H.R. Geminiviruses and Plant Hosts: A Closer Examination of the Molecular Arms Race. Viruses 2017, 9, 256. https://0-doi-org.brum.beds.ac.uk/10.3390/v9090256
Ramesh SV, Sahu PP, Prasad M, Praveen S, Pappu HR. Geminiviruses and Plant Hosts: A Closer Examination of the Molecular Arms Race. Viruses. 2017; 9(9):256. https://0-doi-org.brum.beds.ac.uk/10.3390/v9090256
Chicago/Turabian StyleRamesh, Shunmugiah V., Pranav P. Sahu, Manoj Prasad, Shelly Praveen, and Hanu R. Pappu. 2017. "Geminiviruses and Plant Hosts: A Closer Examination of the Molecular Arms Race" Viruses 9, no. 9: 256. https://0-doi-org.brum.beds.ac.uk/10.3390/v9090256




