mRNA Expression and Activity of Nucleoside Transporters in Human Hepatoma HepaRG Cells
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals and Reagents
2.2. Cell Culture
2.3. RNA Isolation and Analysis
2.4. Immunofluorescence Assays
2.5. Transport Assays
2.6. Statistical Analysis
3. Results
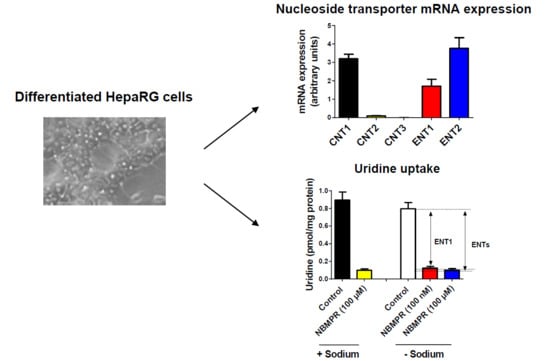
3.1. Expression of Nucleoside Transporters in HepaRG cells
3.2. Nucleoside Transporter Activities in HepaRG Cells
3.3. Regulation of Nucleoside Transporter Expression by Protein Kinase C (PKC) Activation
3.4. Regulation of Nucleoside Transporter Expression by Inflammatory and Growth Factors
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Gripon, P.; Rumin, S.; Urban, S.; Le Seyec, J.; Glaise, D.; Cannie, I.; Guyomard, C.; Lucas, J.; Trepo, C.; Guguen-Guillouzo, C. Infection of a human hepatoma cell line by hepatitis B virus. Proc. Natl. Acad. Sci. USA 2002, 99, 15655–15660. [Google Scholar] [CrossRef] [PubMed]
- Antherieu, S.; Chesne, C.; Li, R.; Guguen-Guillouzo, C.; Guillouzo, A. Optimization of the HepaRG cell model for drug metabolism and toxicity studies. Toxicol. In Vitro 2012, 26, 1278–1285. [Google Scholar] [CrossRef] [PubMed]
- Andersson, T.B.; Kanebratt, K.P.; Kenna, J.G. The HepaRG cell line: A unique in vitro tool for understanding drug metabolism and toxicology in human. Expert. Opin. Drug Metab. Toxicol. 2012, 8, 909–920. [Google Scholar] [CrossRef] [PubMed]
- Aninat, C.; Piton, A.; Glaise, D.; Le Charpentier, T.; Langouet, S.; Morel, F.; Guguen-Guillouzo, C.; Guillouzo, A. Expression of cytochromes P450, conjugating enzymes and nuclear receptors in human hepatoma HepaRG cells. Drug Metab. Dispos. 2006, 34, 75–83. [Google Scholar] [CrossRef] [PubMed]
- Le Vee, M.; Jigorel, E.; Glaise, D.; Gripon, P.; Guguen-Guillouzo, C.; Fardel, O. Functional expression of sinusoidal and canalicular hepatic drug transporters in the differentiated human hepatoma HepaRG cell line. Eur. J. Pharm. Sci. 2006, 28, 109–117. [Google Scholar] [CrossRef] [PubMed]
- Le Vee, M.; Noel, G.; Jouan, E.; Stieger, B.; Fardel, O. Polarized expression of drug transporters in differentiated human hepatoma HepaRG cells. Toxicol. In Vitro 2013, 27, 1979–1986. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Le Vee, M.; Jouan, E.; Noel, G.; Stieger, B.; Fardel, O. Polarized location of SLC and ABC drug transporters in monolayer-cultured human hepatocytes. Toxicol. In Vitro 2015, 29, 938–946. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Molina-Arcas, M.; Casado, F.J.; Pastor-Anglada, M. Nucleoside transporter proteins. Curr. Vasc. Pharmacol. 2009, 7, 426–434. [Google Scholar] [CrossRef] [PubMed]
- Young, J.D.; Yao, S.Y.; Baldwin, J.M.; Cass, C.E.; Baldwin, S.A. The human concentrative and equilibrative nucleoside transporter families, SLC28 and SLC29. Mol. Aspects Med. 2013, 34, 529–547. [Google Scholar] [CrossRef] [PubMed]
- Gray, J.H.; Owen, R.P.; Giacomini, K.M. The concentrative nucleoside transporter family, SLC28. Pflugers Arch. 2004, 447, 728–734. [Google Scholar] [CrossRef] [PubMed]
- Baldwin, S.A.; Beal, P.R.; Yao, S.Y.; King, A.E.; Cass, C.E.; Young, J.D. The equilibrative nucleoside transporter family, SLC29. Pflugers Arch. 2004, 447, 735–743. [Google Scholar] [PubMed]
- Baldwin, S.A.; Yao, S.Y.; Hyde, R.J.; Ng, A.M.; Foppolo, S.; Barnes, K.; Ritzel, M.W.; Cass, C.E.; Young, J.D. Functional characterization of novel human and mouse equilibrative nucleoside transporters (hENT3 and mENT3) located in intracellular membranes. J. Biol. Chem. 2005, 280, 15880–15887. [Google Scholar] [CrossRef] [PubMed]
- Engel, K.; Wang, J. Interaction of organic cations with a newly identified plasma membrane monoamine transporter. Mol. Pharmacol. 2005, 68, 1397–1407. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.; Duan, H.; Engel, K.; Xia, L.; Wang, J. Adenosine transport by plasma membrane monoamine transporter: Reinvestigation and comparison with organic cations. Drug Metab. Dispos. 2010, 38, 1798–1805. [Google Scholar] [CrossRef] [PubMed]
- Pastor-Anglada, M.; Cano-Soldado, P.; Errasti-Murugarren, E.; Casado, F.J. SLC28 genes and concentrative nucleoside transporter (CNT) proteins. Xenobiotica 2008, 38, 972–994. [Google Scholar] [CrossRef] [PubMed]
- Kong, W.; Engel, K.; Wang, J. Mammalian nucleoside transporters. Curr. Drug Metab. 2004, 5, 63–84. [Google Scholar] [CrossRef] [PubMed]
- Pastor-Anglada, M.; Perez-Torras, S. Nucleoside transporter proteins as biomarkers of drug responsiveness and drug targets. Front. Pharmacol. 2015, 6, 13. [Google Scholar] [CrossRef] [PubMed]
- Govindarajan, R.; Bakken, A.H.; Hudkins, K.L.; Lai, Y.; Casado, F.J.; Pastor-Anglada, M.; Tse, C.M.; Hayashi, J.; Unadkat, J.D. In situ hybridization and immunolocalization of concentrative and equilibrative nucleoside transporters in the human intestine, liver, kidneys, and placenta. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2007, 293, R1809–R1822. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Govindarajan, R.; Endres, C.J.; Whittington, D.; LeCluyse, E.; Pastor-Anglada, M.; Tse, C.M.; Unadkat, J.D. Expression and hepatobiliary transport characteristics of the concentrative and equilibrative nucleoside transporters in sandwich-cultured human hepatocytes. Am. J. Physiol. Gastrointest. Liver Physiol. 2008, 295, G570–G580. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fukuchi, Y.; Furihata, T.; Hashizume, M.; Iikura, M.; Chiba, K. Characterization of ribavirin uptake systems in human hepatocytes. J. Hepatol. 2010, 52, 486–492. [Google Scholar] [CrossRef] [PubMed]
- Choi, M.K.; Kim, M.H.; Maeng, H.J.; Song, I.S. Contribution of CNT1 and ENT1 to ribavirin uptake in human hepatocytes. Arch. Pharm. Res. 2015, 38, 904–913. [Google Scholar] [CrossRef] [PubMed]
- Shimada, T.; Nakanishi, T.; Tajima, H.; Yamazaki, M.; Yokono, R.; Takabayashi, M.; Sawamoto, K.; Miyamoto, K.; Kitagawa, H.; Ohta, T.; et al. Saturable Hepatic Extraction of Gemcitabine Involves Biphasic Uptake Mediated by Nucleoside Transporters Equilibrative Nucleoside Transporter 1 and 2. J. Pharm. Sci. 2015, 104, 3162–3169. [Google Scholar] [CrossRef] [PubMed]
- Mayati, A.; Le Vee, M.; Moreau, A.; Jouan, E.; Bucher, S.; Stieger, B.; Denizot, C.; Parmentier, Y.; Fardel, O. Protein kinase C-dependent regulation of human hepatic drug transporter expression. Biochem. Pharmacol. 2015, 98, 703–717. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fardel, O.; Le Vee, M. Regulation of human hepatic drug transporter expression by pro-inflammatory cytokines. Expert Opin. Drug Metab. Toxicol. 2009, 5, 1469–1481. [Google Scholar] [CrossRef] [PubMed]
- Jouan, E.; Le Vee, M.; Denizot, C.; Parmentier, Y.; Fardel, O. Drug Transporter Expression and Activity in Human Hepatoma HuH-7 Cells. Pharmaceutics 2016, 9, 3. [Google Scholar] [CrossRef] [PubMed]
- Van Grevenynghe, J.; Rion, S.; Le Ferrec, E.; Le Vee, M.; Amiot, L.; Fauchet, R.; Fardel, O. Polycyclic aromatic hydrocarbons inhibit differentiation of human monocytes into macrophages. J. Immunol. 2003, 170, 2374–2381. [Google Scholar] [CrossRef] [PubMed]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef] [PubMed]
- Le Vee, M.; Gripon, P.; Stieger, B.; Fardel, O. Down-regulation of organic anion transporter expression in human hepatocytes exposed to the proinflammatory cytokine interleukin 1beta. Drug Metab. Dispos. 2008, 36, 217–222. [Google Scholar] [CrossRef] [PubMed]
- Mayati, A.; Moreau, A.; Denizot, C.; Stieger, B.; Parmentier, Y.; Fardel, O. beta2-adrenergic receptor-mediated in vitro regulation of human hepatic drug transporter expression by epinephrine. Eur. J. Pharm. Sci. 2017, 106, 302–312. [Google Scholar] [CrossRef] [PubMed]
- Klein, D.M.; Evans, K.K.; Hardwick, R.N.; Dantzler, W.H.; Wright, S.H.; Cherrington, N.J. Basolateral uptake of nucleosides by Sertoli cells is mediated primarily by equilibrative nucleoside transporter 1. J. Pharmacol. Exp. Ther. 2013, 346, 121–129. [Google Scholar] [CrossRef] [PubMed]
- Furihata, T.; Fukuchi, Y.; Iikura, M.; Hashizume, M.; Miyajima, A.; Nagai, M.; Chiba, K. Striking species difference in the contribution of concentrative nucleoside transporter 2 to nucleoside uptake between mouse and rat hepatocytes. Antimicrob. Agents Chemother. 2010, 54, 3035–3038. [Google Scholar] [CrossRef] [PubMed]
- Bachour-El Azzi, P.; Sharanek, A.; Burban, A.; Li, R.; Guevel, R.L.; Abdel-Razzak, Z.; Stieger, B.; Guguen-Guillouzo, C.; Guillouzo, A. Comparative Localization and Functional Activity of the Main Hepatobiliary Transporters in HepaRG Cells and Primary Human Hepatocytes. Toxicol. Sci. 2015, 145, 157–168. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mayati, A.; Moreau, A.; Le Vee, M.; Stieger, B.; Denizot, C.; Parmentier, Y.; Fardel, O. Protein Kinases C-Mediated Regulations of Drug Transporter Activity, Localization and Expression. Int. J. Mol. Sci. 2017, 18, 764. [Google Scholar] [CrossRef] [PubMed]
- Toullec, D.; Pianetti, P.; Coste, H.; Bellevergue, P.; Grand-Perret, T.; Ajakane, M.; Baudet, V.; Boissin, P.; Boursier, E.; Loriolle, F.; et al. The bisindolylmaleimide GF 109203X is a potent and selective inhibitor of protein kinase C. J. Biol. Chem. 1991, 266, 15771–15781. [Google Scholar] [PubMed]
- Martiny-Baron, G.; Kazanietz, M.G.; Mischak, H.; Blumberg, P.M.; Kochs, G.; Hug, H.; Marme, D.; Schachtele, C. Selective inhibition of protein kinase C isozymes by the indolocarbazole Go 6976. J. Biol. Chem. 1993, 268, 9194–9197. [Google Scholar] [PubMed]
- Le Vee, M.; Lecureur, V.; Moreau, A.; Stieger, B.; Fardel, O. Differential regulation of drug transporter expression by hepatocyte growth factor in primary human hepatocytes. Drug Metab. Dispos. 2009, 37, 2228–2235. [Google Scholar] [CrossRef] [PubMed]
- Febvre-James, M.; Bruyere, A.; Le Vee, M.; Fardel, O. The JAK1/2 Inhibitor Ruxolitinib Reverses Interleukin-6-Mediated Suppression of Drug-Detoxifying Proteins in Cultured Human Hepatocytes. Drug Metab. Dispos. 2018, 46, 131–140. [Google Scholar] [CrossRef] [PubMed]
- Guillouzo, A.; Corlu, A.; Aninat, C.; Glaise, D.; Morel, F.; Guguen-Guillouzo, C. The human hepatoma HepaRG cells: A highly differentiated model for studies of liver metabolism and toxicity of xenobiotics. Chem. Biol. Interact. 2007, 168, 66–73. [Google Scholar] [CrossRef] [PubMed]
- Pennycooke, M.; Chaudary, N.; Shuralyova, I.; Zhang, Y.; Coe, I.R. Differential expression of human nucleoside transporters in normal and tumor tissue. Biochem. Biophys. Res. Commun. 2001, 280, 951–959. [Google Scholar] [CrossRef] [PubMed]
- Ritzel, M.W.; Yao, S.Y.; Ng, A.M.; Mackey, J.R.; Cass, C.E.; Young, J.D. Molecular cloning, functional expression and chromosomal localization of a cDNA encoding a human Na+/nucleoside cotransporter (hCNT2) selective for purine nucleosides and uridine. Mol. Membr. Biol. 1998, 15, 203–211. [Google Scholar] [PubMed]
- Mele, M.; Ferreira, P.G.; Reverter, F.; DeLuca, D.S.; Monlong, J.; Sammeth, M.; Young, T.R.; Goldmann, J.M.; Pervouchine, D.D.; Sullivan, T.J.; et al. Human genomics. The human transcriptome across tissues and individuals. Science 2015, 348, 660–665. [Google Scholar] [CrossRef] [PubMed]
- Ritzel, M.W.; Ng, A.M.; Yao, S.Y.; Graham, K.; Loewen, S.K.; Smith, K.M.; Ritzel, R.G.; Mowles, D.A.; Carpenter, P.; Chen, X.Z.; et al. Molecular identification and characterization of novel human and mouse concentrative Na+-nucleoside cotransporter proteins (hCNT3 and mCNT3) broadly selective for purine and pyrimidine nucleosides (system cib). J. Biol. Chem. 2001, 276, 2914–2927. [Google Scholar] [CrossRef] [PubMed]
- Hansen, J.E.; Tse, C.M.; Chan, G.; Heinze, E.R.; Nishimura, R.N.; Weisbart, R.H. Intranuclear protein transduction through a nucleoside salvage pathway. J. Biol. Chem. 2007, 282, 20790–20793. [Google Scholar] [CrossRef] [PubMed]
- Klein, D.M.; Harding, M.C.; Crowther, M.K.; Cherrington, N.J. Localization of nucleoside transporters in rat epididymis. J. Biochem. Mol. Toxicol. 2017, 31, e21911. [Google Scholar] [CrossRef] [PubMed]
- Grane-Boladeras, N.; Spring, C.M.; Hanna, W.J.; Pastor-Anglada, M.; Coe, I.R. Novel nuclear hENT2 isoforms regulate cell cycle progression via controlling nucleoside transport and nuclear reservoir. Cell. Mol. Life Sci. 2016, 73, 4559–4575. [Google Scholar] [CrossRef] [PubMed]
- Burns, D.J.; Bell, R.M. Protein kinase C contains two phorbol ester binding domains. J. Biol. Chem. 1991, 266, 18330–18338. [Google Scholar] [PubMed]
- Hartmann, G.; Kim, H.; Piquette-Miller, M. Regulation of the hepatic multidrug resistance gene expression by endotoxin and inflammatory cytokines in mice. Int. Immunopharmacol. 2001, 1, 189–199. [Google Scholar] [CrossRef]
- Diao, L.; Li, N.; Brayman, T.G.; Hotz, K.J.; Lai, Y. Regulation of MRP2/ABCC2 and BSEP/ABCB11 expression in sandwich cultured human and rat hepatocytes exposed to inflammatory cytokines TNF-α, IL-6, and IL-1β. J. Biol. Chem. 2010, 285, 31185–31192. [Google Scholar] [CrossRef] [PubMed]
- Aitken, A.E.; Richardson, T.A.; Morgan, E.T. Regulation of drug-metabolizing enzymes and transporters in inflammation. Annu. Rev. Pharmacol. Toxicol. 2006, 46, 123–149. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Veledo, S.; Valdes, R.; Wallenius, V.; Casado, F.J.; Pastor-Anglada, M. Up-regulation of the high-affinity pyrimidine-preferring nucleoside transporter concentrative nucleoside transporter 1 by tumor necrosis factor-alpha and interleukin-6 in liver parenchymal cells. J. Hepatol. 2004, 41, 538–544. [Google Scholar] [CrossRef] [PubMed]
- Soler, C.; Felipe, A.; Garcia-Manteiga, J.; Serra, M.; Guillen-Gomez, E.; Casado, F.J.; MacLeod, C.; Modolell, M.; Pastor-Anglada, M.; Celada, A. Interferon-gamma regulates nucleoside transport systems in macrophages through signal transduction and activator of transduction factor 1 (STAT1)-dependent and -independent signalling pathways. Biochem. J. 2003, 375, 777–783. [Google Scholar] [CrossRef] [PubMed]
- Ezura, Y.; Lin, X.; Hatta, A.; Izu, Y.; Noda, M. Interleukin-1beta Suppresses the Transporter Genes Ank and Ent1 Expression in Stromal Progenitor Cells Retaining Mineralization. Calcif. Tissue Int. 2016, 99, 199–208. [Google Scholar] [CrossRef] [PubMed]
- Pinilla-Macua, I.; Fernandez-Calotti, P.; Perez-Del-Pulgar, S.; Pastor-Anglada, M. Ribavirin uptake into human hepatocyte HHL5 cells is enhanced by interferon-alpha via up-regulation of the human concentrative nucleoside transporter (hCNT2). Mol. Pharm. 2014, 11, 3223–3230. [Google Scholar] [CrossRef] [PubMed]
- Baloch, K.; Chen, L.; Memon, A.A.; Dexter, L.; Irving, W.; Ilyas, M.; Thomson, B.J. Equilibrative nucleoside transporter 1 expression in primary human hepatocytes is highly variable and determines uptake of ribavirin. Antivir. Chem. Chemother. 2017, 25, 2–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]










| Inhibitor | Nucleoside Transporter | ||||
|---|---|---|---|---|---|
| CNT1 | CNT2 | CNT3 | ENT1 | ENT2 | |
| Sodium whithdrawal | + | + | + | − | − |
| S-(4-Nitrobenzyl)-6-thioinosine (NBMPR) (100 nM) | − | − | − | + | − |
| NBMPR (100 µM) | − | − | − | + | + |
| Inosine (200 µM) | − | + | + | Not relevant | Not relevant |
| Thymidine (200 µM) | + | − | + | Not relevant | Not relevant |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mayati, A.; Moreau, A.; Jouan, E.; Febvre-James, M.; Denizot, C.; Parmentier, Y.; Fardel, O. mRNA Expression and Activity of Nucleoside Transporters in Human Hepatoma HepaRG Cells. Pharmaceutics 2018, 10, 246. https://0-doi-org.brum.beds.ac.uk/10.3390/pharmaceutics10040246
Mayati A, Moreau A, Jouan E, Febvre-James M, Denizot C, Parmentier Y, Fardel O. mRNA Expression and Activity of Nucleoside Transporters in Human Hepatoma HepaRG Cells. Pharmaceutics. 2018; 10(4):246. https://0-doi-org.brum.beds.ac.uk/10.3390/pharmaceutics10040246
Chicago/Turabian StyleMayati, Abdullah, Amélie Moreau, Elodie Jouan, Marie Febvre-James, Claire Denizot, Yannick Parmentier, and Olivier Fardel. 2018. "mRNA Expression and Activity of Nucleoside Transporters in Human Hepatoma HepaRG Cells" Pharmaceutics 10, no. 4: 246. https://0-doi-org.brum.beds.ac.uk/10.3390/pharmaceutics10040246