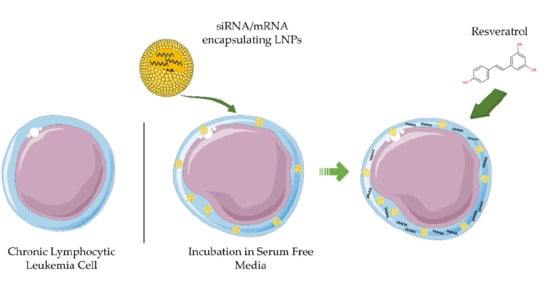
Resveratrol Enhances mRNA and siRNA Lipid Nanoparticles Primary CLL Cell Transfection
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. LNP Preparation and Characterization
2.3. Transfection Protocol
2.4. Separation of CLL Cells from Patient Samples
2.5. LNP Uptake Experiments
2.6. Confocal Microscopy Analysis
2.7. Luciferase Expression
2.8. Mcl-1, CD44, STAT3 Silencing, and Cell Viability Assessment
- siMcl-1 sense: CCCGCCGAAUUCAUUAAUUUACUGT,
- anti-sense: ACAGUAAAUUAAUGAAUUCGGCGGGUA;
- siCD44 sense: GGCGCAGAUCGAUUUGAAUAUAACC,
- anti-sense: GGUUAUAUUCAAAUCGAUCUGCGCCCAG;
- siSTAT3 sense: CAGCAACACUCUUCAGUACAUAAUA,
- anti-sense: UAUUAUGUACUGAAGAGUGUUGCUGGA;
- siNC5 sense: CAUAUUGCGCGUAUAGUCGCGUUAG,
- anti-sense: UGGUAUAACGCGCAUAUCAGCGAAUC.
2.9. RT-PCR Experiments
3. Results
3.1. Physicochemical and Structural Characterization of LNPs
3.2. Serum Depletion Results in Rapid LNP Uptake into CLL Cells which Enhances mRNA-LNPs Transfection
3.3. Resveratrol Enhances mRNA LNP Transfection into Primary CLL Cells
3.4. Resveratrol Enhances siRNA LNP Transfection into Primary CLL Cells
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Pulte, D.; Castro, F.A.; Jansen, L.; Luttmann, S.; Holleczek, B.; Nennecke, A.; Ressing, M.; Katalinic, A.; Brenner, H. Trends in survival of chronic lymphocytic leukemia patients in Germany and the USA in the first decade of the twenty-first century. J. Hematol. Oncol. 2016, 9, 28. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Delgado, J.; Doubek, M.; Baumann, T.; Kotaskova, J.; Molica, S.; Mozas, P.; Rivas-Delgado, A.; Morabito, F.; Pospisilova, S.; Montserrat, E. Chronic lymphocytic leukemia: A prognostic model comprising only two biomarkers (IGHV mutational status and FISH cytogenetics) separates patients with different outcome and simplifies the CLL-IPI. Am. J. Hematol. 2017, 92, 375–380. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nabhan, C.; Rosen, S.T. Chronic Lymphocytic Leukemia: A Clinical Review. JAMA J. Am. Med. Assoc. 2014, 312, 2265–2276. [Google Scholar] [CrossRef] [Green Version]
- Burger, J.A.; O’Brien, S. Evolution of CLL treatment—From chemoimmunotherapy to targeted and individualized therapy. Nat. Rev. Clin. Oncol. 2018, 15, 510–527. [Google Scholar] [CrossRef] [PubMed]
- Seiffert, M.; Stilgenbauer, S.; Döhner, H.; Lichter, P. Efficient nucleofection of primary human B cells and B-CLL cells induces apoptosis, which depends on the microenvironment and on the structure of transfected nucleic acids. Leukemia 2007, 21, 1977–1983. [Google Scholar] [CrossRef]
- Matutes, E.; Owusu-Ankomah, K.; Morilla, R.; Garcia Marco, J.; Houlihan, A.; Que, T.H.; Catovsky, D. The immunological profile of B-cell disorders and proposal of a scoring system for the diagnosis of CLL. Leukemia 1994, 8, 1640–1645. [Google Scholar]
- Wendtner, C.-M.; Gregor, M. Current perspectives on the role of chemotherapy in chronic lymphocytic leukemia. Leuk. Lymphoma 2018, 59, 300–310. [Google Scholar] [CrossRef]
- Pastor, F.; Berraondo, P.; Etxeberria, I.; Frederick, J.; Sahin, U.; Gilboa, E.; Melero, I. An RNA toolbox for cancer immunotherapy. Nat. Rev. Drug Discov. 2018, 17, 751–767. [Google Scholar] [CrossRef]
- Granot-Matok, Y.; Kon, E.; Dammes, N.; Mechtinger, G.; Peer, D. Therapeutic mRNA delivery to leukocytes. J. Control. Release 2019, 305, 165–175. [Google Scholar] [CrossRef]
- Urquhart, L. FDA new drug approvals in Q3 2018. Nat. Rev. Drug Discov. 2018, 17, 779. [Google Scholar] [CrossRef]
- Adams, D.; Gonzalez-Duarte, A.; O’Riordan, W.D.; Yang, C.-C.; Ueda, M.; Kristen, A.V.; Tournev, I.; Schmidt, H.H.; Coelho, T.; Berk, J.L.; et al. Patisiran, an RNAi Therapeutic, for Hereditary Transthyretin Amyloidosis. N. Eng. J. Med. 2018, 379, 11–21. [Google Scholar] [CrossRef] [PubMed]
- Leung, A.K.K.; Tam, Y.Y.C.; Cullis, P.R. Lipid nanoparticles for short Interfering RNA delivery. Adv. Genet. 2014, 88, 71–110. [Google Scholar] [PubMed]
- Durymanov, M.; Reineke, J. Non-viral Delivery of Nucleic Acids: Insight Into Mechanisms of Overcoming Intracellular Barriers. Front. Pharmacol. 2018, 9, 1–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Healey, D.G.; Ketteringham, H.; Frandji, P.; Simon, J.; Amir, A.; Harris, S.J.; Mountain, A.; Alistair, S.; Fdq, Z.; Eh, R.Q.O.; et al. 354. Efficient Non-Viral Transfection of CLL-B Cells with Human CD40 Ligand. Mol. Ther. 2002, 5, S117. [Google Scholar]
- Tam, Y.Y.C.; Chen, S.; Cullis, P.R. Advances in lipid nanoparticles for siRNA delivery. Pharmaceutics 2013, 5, 498–507. [Google Scholar] [CrossRef] [Green Version]
- Kulkarni, J.A.; Darjuan, M.M.; Mercer, J.E.; Chen, S.; Van Der Meel, R.; Thewalt, J.L.; Tam, Y.Y.C.; Cullis, P.R. On the Formation and Morphology of Lipid Nanoparticles Containing Ionizable Cationic Lipids and siRNA. ACS Nano 2018, 12, 4787–4795. [Google Scholar] [CrossRef] [Green Version]
- Rosenblum, D.; Joshi, N.; Tao, W.; Karp, J.M. Dan Peer Progress and challenges towards targeted delivery of cancer therapeutics. Nat. Commun. 2018, 9, 1410. [Google Scholar] [CrossRef] [Green Version]
- Vermeulen, L.M.P.; Brans, T.; Samal, S.K.; Dubruel, P.; Demeester, J.; De Smedt, S.C.; Remaut, K.; Braeckmans, K. Endosomal Size and Membrane Leakiness Influence Proton Sponge-Based Rupture of Endosomal Vesicles. ACS Nano 2018, 12, 2332–2345. [Google Scholar] [CrossRef] [Green Version]
- Abbas, M.; Saeed, F.; Anjum, F.M.; Afzaal, M.; Tufail, T.; Bashir, M.S.; Ishtiaq, A.; Hussain, S.; Suleria, H.A.R. Natural polyphenols: An overview. Int. J. Food Prop. 2017, 20, 1689–1699. [Google Scholar] [CrossRef] [Green Version]
- Ko, J.H.; Sethi, G.; Um, J.Y.; Shanmugam, M.K.; Arfuso, F.; Kumar, A.P.; Bishayee, A.; Ahn, K.S. The role of resveratrol in cancer therapy. Int. J. Mol. Sci. 2017, 18, 2589. [Google Scholar] [CrossRef] [Green Version]
- Pelinson, L.P.; Assmann, C.E.; Palma, T.V.; da Cruz, I.B.M.; Pillat, M.M.; Mânica, A.; Stefanello, N.; Weis, G.C.C.; de Oliveira Alves, A.; de Andrade, C.M.; et al. Antiproliferative and apoptotic effects of caffeic acid on SK-Mel-28 human melanoma cancer cells. Mol. Biol. Rep. 2019, 46, 2085–2092. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.; Zhang, S.; Shen, H.; Chen, W.; Xu, H.; Chen, X.; Sun, D.; Zhong, S.; Zhao, J.; Tang, J. Curcumin inhibits cancer progression through regulating expression of microRNAs. Tumor Biol. 2017, 39, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jang, M.; Jang, M.; Cai, L.; Udeani, G.O.; Slowing, K.V.; Thomas, C.F.; Beecher, C.W.W.; Fong, H.H.S.; Farnsworth, N.R.; Kinghorn, A.D.; et al. Cancer Chemopreventive Activity of Resveratrol, a Natural Product Derived from Grapes Cancer. Science 1997, 275, 218–220. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baur, J.A.; Sinclair, D.A. Therapeutic potential of resveratrol: The in vivo evidence. Nat. Rev. Drug Discov. 2006, 5, 493–506. [Google Scholar] [CrossRef]
- Rauf, A.; Imran, M.; Butt, M.S.; Nadeem, M.; Peters, D.G.; Mubarak, M.S. Resveratrol as an anti-cancer agent: A review. Crit. Rev. Food Sci. Nutr. 2018, 58, 1428–1447. [Google Scholar] [CrossRef]
- Garber, K. Alnylam launches era of RNAi drugs. Nat. Biotechnol. 2018, 36, 777–778. [Google Scholar] [CrossRef]
- Kowalski, P.S.; Rudra, A.; Miao, L.; Anderson, D.G. Delivering the Messenger: Advances in Technologies for Therapeutic mRNA Delivery. Mol. Ther. 2019, 27, 710–728. [Google Scholar] [CrossRef] [Green Version]
- Kulkarni, J.A.; Witzigmann, D.; Chen, S.; Cullis, P.R.; van der Meel, R. Lipid Nanoparticle Technology for Clinical Translation of siRNA Therapeutics. Acc. Chem. Res. 2019, 9, 2435–2444. [Google Scholar] [CrossRef]
- Du Rietz, H.; Hedlund, H.; Wilhelmson, S.; Nordenfelt, P.; Wittrup, A. Imaging small molecule-induced endosomal escape of siRNA. Nat. Commun. 2020, 11, 1–17. [Google Scholar] [CrossRef]
- Lönn, P.; Kacsinta, A.D.; Cui, X.S.; Hamil, A.S.; Kaulich, M.; Gogoi, K.; Dowdy, S.F. Enhancing Endosomal Escape for Intracellular Delivery of Macromolecular Biologic Therapeutics. Sci. Rep. 2016, 6, 1–9. [Google Scholar] [CrossRef]
- Lv, H.; Zhang, S.; Wang, B.; Cui, S.; Yan, J. Toxicity of cationic lipids and cationic polymers in gene delivery. J. Control. Release 2006, 114, 100–109. [Google Scholar] [CrossRef] [PubMed]
- Johnston, J.B.; Paul, J.T.; Neufeld, N.J.; Haney, N.; Kropp, D.M.; Hu, X.; Cheang, M.; Gibson, S.B. Role of myeloid cell factor-1 (Mcl-1) in chronic lymphocytic leukemia. Leuk. Lymphoma 2004, 45, 2017–2027. [Google Scholar] [CrossRef] [PubMed]
- Pepper, C.; Lin, T.T.; Pratt, G.; Hewamana, S.; Brennan, P.; Hiller, L.; Hills, R.; Ward, R.; Starczynski, J.; Austen, B.; et al. Mcl-1 expression has in vitro and in vivo significance in chronic lymphocytic leukemia and is associated with other poor prognostic markers. J. Am. Soc. Hematol. 2015, 112, 3807–3818. [Google Scholar] [CrossRef]
- Kotschy, A.; Szlavik, Z.; Murray, J.; Davidson, J.; Maragno, A.L.; Le Toumelin-Braizat, G.; Chanrion, M.; Kelly, G.L.; Gong, J.N.; Moujalled, D.M.; et al. The MCL1 inhibitor S63845 is tolerable and effective in diverse cancer models. Nature 2016, 538, 477–482. [Google Scholar] [CrossRef] [PubMed]
- Kondo, K.; Shaim, H.; Thompson, P.A.; Burger, J.A.; Keating, M.; Estrov, Z.; Harris, D.; Kim, E.; Ferrajoli, A.; Daher, M.; et al. Ibrutinib modulates the immunosuppressive CLL microenvironment through STAT3-mediated suppression of regulatory B-cell function and inhibition of the PD-1/PD-L1 pathway. Leukemia 2018, 32, 960–970. [Google Scholar] [CrossRef] [PubMed]
- Lin, F.; Wu, D.; Fang, D.; Chen, Y.; Zhou, H.; Ou, C. STAT3-induced SMYD3 transcription enhances chronic lymphocytic leukemia cell growth in vitro and in vivo. Inflamm. Res. 2019, 68, 739–749. [Google Scholar] [CrossRef]
- Hazan-Halevy, I.; Harris, D.; Liu, Z.; Liu, J.; Li, P.; Chen, X.; Shanker, S.; Ferrajoli, A.; Keating, M.J.; Estrov, Z. STAT3 is constitutively phosphorylated on serine 727 residues, binds DNA, and activates transcription in CLL cells. Blood 2010, 115, 2852–2863. [Google Scholar] [CrossRef] [Green Version]
- Gutjahr, J.C.; Greil, R.; Hartmann, T.N. The role of CD44 in the pathophysiology of chronic lymphocytic leukemia. Front. Immunol. 2015, 6, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Morath, I.; Hartmann, T.N.; Orian-Rousseau, V. CD44: More than a mere stem cell marker. Int. J. Biochem. Cell Biol. 2016, 81, 166–173. [Google Scholar] [CrossRef]
- Zheng, C.Y.; Paui, P.; Maksymiuk, A.W.; Skinnider, L.F. Establishment of cell lines derived from chronic lymphocytic leukaemic cells by transfection with myc and ras. Br. J. Haematol. 1996, 93, 681–683. [Google Scholar] [CrossRef]
- Kulkarni, J.A.; Cullis, P.R.; Van Der Meel, R. Lipid Nanoparticles Enabling Gene Therapies: From Concepts to Clinical Utility. Nucleic Acid Ther. 2018, 28, 146–157. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, A.-M.; Tsunoda, I.; Yoshie, O. Heat shock protein 27 promotes cell cycle progression by down-regulating E2F transcription factor 4 and retinoblastoma family protein p130. J. Biol. Chem. 2018, 293, 15815–15826. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiong, W.; Jiao, Y.; Huang, W.; Ma, M.; Yu, M.; Cui, Q.; Tan, D. Regulation of the cell cycle via mitochondrial gene expression and energy metabolism in HeLa cells. Acta Biochim. Biophys. Sin. 2012, 44, 347–358. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, D.; Ganesh, S.; Wang, W.; Amiji, M. The role of surface chemistry in serum protein corona-mediated cellular delivery and gene silencing with lipid nanoparticles. Nanoscale 2019, 11, 8760–8775. [Google Scholar] [CrossRef]
- Akinc, A.; Querbes, W.; De, S.; Qin, J.; Frank-Kamenetsky, M.; Jayaprakash, K.N.; Jayaraman, M.; Rajeev, K.G.; Cantley, W.L.; Dorkin, J.R.; et al. Targeted delivery of RNAi therapeutics with endogenous and exogenous ligand-based mechanisms. Mol. Ther. 2010, 18, 1357–1364. [Google Scholar] [CrossRef]
- Maugeri, M.; Nawaz, M.; Papadimitriou, A.; Angerfors, A.; Camponeschi, A.; Na, M.; Hölttä, M.; Skantze, P.; Johansson, S.; Sundqvist, M.; et al. Linkage between endosomal escape of LNP-mRNA and loading into EVs for transport to other cells. Nat. Commun. 2019, 10. [Google Scholar] [CrossRef] [Green Version]
- Tomic, J.; McCaw, L.; Li, Y.; Hough, M.R.; Ben-David, Y.; Moffat, J.; Spaner, D.E. Resveratrol has anti-leukemic activity associated with decreased O-GlcNAcylated proteins. Exp. Hematol. 2013, 41, 675–686. [Google Scholar] [CrossRef]
- Podhorecka, M.; Halicka, D.; Klimek, P.; Kowal, M.; Chocholska, S.; Dmoszynska, A. Resveratrol increases rate of apoptosis caused by purine analogues in malignant lymphocytes of chronic lymphocytic leukemia. Ann. Hematol. 2011, 90, 1–15, 173–183. [Google Scholar] [CrossRef] [Green Version]
- Gokbulut, A.A.; Apohan, E.; Baran, Y. Resveratrol and quercetin-induced apoptosis of human 232B4 chronic lymphocytic leukemia cells by activation of caspase-3 and cell cycle arrest. Hematology 2013, 18, 144–150. [Google Scholar] [CrossRef] [Green Version]
- Jhaveri, A.; Deshpande, P.; Pattni, B.; Torchilin, V. Transferrin-targeted, resveratrol-loaded liposomes for the treatment of glioblastoma. J. Control. Release 2018, 277, 89–101. [Google Scholar] [CrossRef]
- Espinoza, J.L.; Kurokawa, Y.; Takami, A. Rationale for assessing the therapeutic potential of resveratrol in hematological malignancies. Blood Rev. 2019, 33, 43–52. [Google Scholar] [CrossRef] [PubMed]
- Gautam, S.C.; Xu, Y.X.; Dumaguin, M.; Janakiraman, N.; Chapman, R.A. Resveratrol selectively inhibits leukemia cells: A prospective agent for ex vivo bone marrow purging. Bone Marrow Transplant. 2000, 25, 639–645. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kedmi, R.; Veiga, N.; Ramishetti, S.; Goldsmith, M.; Rosenblum, D.; leviatan-ben arye, S.; Harlev, M.; Benhar, I.; Lieberman, J.; Peer, D. A modular platform for targeted RNAi therapeutics. Nat. Nanotechnol. 2018, 13, 214–219. [Google Scholar] [CrossRef] [PubMed]
- Weinstein, S.; Toker, I.A.; Emmanuel, R.; Ramishetti, S.; Hazan-halevy, I. Harnessing RNAi-based nanomedicines for therapeutic gene silencing in B-cell malignancies. Proc. Natl. Acad. Sci. USA 2016, 113, E16–E22. [Google Scholar] [CrossRef] [PubMed] [Green Version]




© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kon, E.; Hazan-Halevy, I.; Rosenblum, D.; Cohen, N.; Chatterjee, S.; Veiga, N.; Raanani, P.; Bairey, O.; Benjamini, O.; Nagler, A.; et al. Resveratrol Enhances mRNA and siRNA Lipid Nanoparticles Primary CLL Cell Transfection. Pharmaceutics 2020, 12, 520. https://0-doi-org.brum.beds.ac.uk/10.3390/pharmaceutics12060520
Kon E, Hazan-Halevy I, Rosenblum D, Cohen N, Chatterjee S, Veiga N, Raanani P, Bairey O, Benjamini O, Nagler A, et al. Resveratrol Enhances mRNA and siRNA Lipid Nanoparticles Primary CLL Cell Transfection. Pharmaceutics. 2020; 12(6):520. https://0-doi-org.brum.beds.ac.uk/10.3390/pharmaceutics12060520
Chicago/Turabian StyleKon, Edo, Inbal Hazan-Halevy, Daniel Rosenblum, Niv Cohen, Sushmita Chatterjee, Nuphar Veiga, Pia Raanani, Osnat Bairey, Ohad Benjamini, Arnon Nagler, and et al. 2020. "Resveratrol Enhances mRNA and siRNA Lipid Nanoparticles Primary CLL Cell Transfection" Pharmaceutics 12, no. 6: 520. https://0-doi-org.brum.beds.ac.uk/10.3390/pharmaceutics12060520