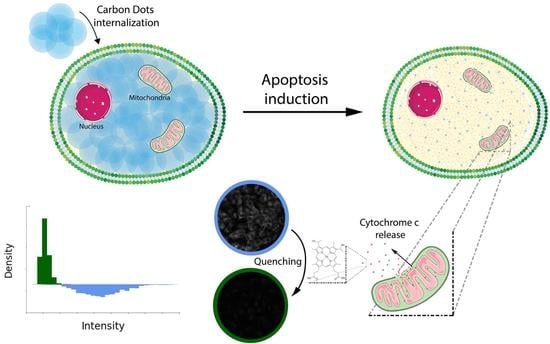
Quantifying Cytosolic Cytochrome c Concentration Using Carbon Quantum Dots as a Powerful Method for Apoptosis Detection
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Carbon Quantum Dots Synthesis
2.3. Optical Properties
2.3.1. UV-Vis Spectroscopy
2.3.2. Transmission Electron Microscopy
2.3.3. The Fourier Transform Infrared (FTIR)
2.3.4. Raman Spectroscopy
2.4. Biological Characterization and In Vitro Studies
2.4.1. Cell Culture
2.4.2. Cell Viability Assays
2.4.3. Confocal Microscopy–Morphological Characterization
2.4.4. Semiquantitative Analysis–Confocal
3. Results and Discussion
3.1. Preparation and Characterization of CQDs
3.2. Optical Properties
3.3. Transmission Electron Microscopy (TEM)
3.4. Fourier Transform Infra-Red Spectroscopy (FTIR)
3.5. Raman Spectroscopy
3.6. In Vitro Cytotoxicity Assessments of the CQDs Solution
3.7. Cellular Uptake
3.8. Confocal Microscopy—Morphological Characterization
3.9. Semiquantitative Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Tait, S.W.G.; Green, D.R. Mitochondria and cell death: Outer membrane permeabilization and beyond. Nat. Rev. Mol. Cell Biol. 2010, 11, 621–632. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zhang, B.; Di, C.; Ali, M.C.; Chen, J.; Li, Z.; Si, J.; Zhang, H.; Qiu, H. Label-free fluorescence imaging of cytochrome c in living systems and anti-cancer drug screening with nitrogen doped carbon quantum dots. Nanoscale 2018, 10, 5342–5349. [Google Scholar] [CrossRef] [PubMed]
- Chandel, N.S. Mitochondria as signaling organelles. BMC Biol. 2014, 12, 34. [Google Scholar] [CrossRef] [PubMed]
- Kroemer, G.; Galluzzi, L.; Brenner, C. Mitochondrial Membrane Permeabilization in Cell Death. Physiol. Rev. 2007, 87, 99–163. [Google Scholar] [CrossRef] [PubMed]
- Garrido, C.; Galluzzi, L.; Brunet, M.; Puig, P.E.; Didelot, C.; Kroemer, G. Mechanisms of cytochrome c release from mitochondria. Cell Death Differ. 2006, 13, 1423–1433. [Google Scholar] [CrossRef] [Green Version]
- Campos, C.B.L.; Paim, B.A.; Cosso, R.G.; Castilho, R.F.; Rottenberg, H.; Vercesi, A.E. Method for monitoring of mitochondrial cytochrome c release during cell death: Immunodetection of cytochrome c by flow cytometry after selective permeabilization of the plasma membrane. Cytom. Part A 2006, 69A, 515–523. [Google Scholar] [CrossRef]
- Waterhouse, N.J.; Trapani, J.A. A new quantitative assay for cytochrome c release in apoptotic cells. Cell Death Differ. 2003, 10, 853–855. [Google Scholar] [CrossRef] [Green Version]
- Amin, R.M.; Elfeky, S.A.; Verwanger, T.; Krammer, B. Fluorescence-based CdTe nanosensor for sensitive detection of cytochrome C. Biosens. Bioelectron. 2017, 98, 415–420. [Google Scholar] [CrossRef]
- Jin, M.; Liu, X.; Zhang, X.; Wang, L.; Bing, T.; Zhang, N.; Zhang, Y.; Shangguan, D. Thiazole Orange-Modified Carbon Dots for Ratiometric Fluorescence Detection of G-Quadruplex and Double-Stranded DNA. ACS Appl. Mater. Interfaces 2018, 10, 25166–25173. [Google Scholar] [CrossRef]
- Wang, J.; Qiu, J. A review of carbon dots in biological applications. J. Mater. Sci. 2016, 51, 4728–4738. [Google Scholar] [CrossRef]
- Das, P.; Maity, P.P.; Ganguly, S.; Ghosh, S.; Baral, J.; Bose, M.; Choudhary, S.; Gangopadhyay, S.; Dhara, S.; Das, A.K.; et al. Biocompatible carbon dots derived from κ-carrageenan and phenyl boronic acid for dual modality sensing platform of sugar and its anti-diabetic drug release behavior. Int. J. Biol. Macromol. 2019, 132, 316–329. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.; Khan, R.; Catanante, G.; Sherazi, T.; Bhand, S.; Hayat, A.; Marty, J. Designed Strategies for Fluorescence-Based Biosensors for the Detection of Mycotoxins. Toxins 2018, 10, 197. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoo, D.; Park, Y.; Cheon, B.; Park, M.-H. Carbon Dots as an Effective Fluorescent Sensing Platform for Metal Ion Detection. Nanoscale Res. Lett. 2019, 14, 272. [Google Scholar] [CrossRef] [Green Version]
- Zarei-Ghobadi, M.; Mozhgani, S.-H.; Dashtestani, F.; Yadegari, A.; Hakimian, F.; Norouzi, M.; Ghourchian, H. A genosensor for detection of HTLV-I based on photoluminescence quenching of fluorescent carbon dots in presence of iron magnetic nanoparticle-capped Au. Sci. Rep. 2018, 8, 15593. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Lu, K.-Q.; Tang, Z.-R.; Xu, Y.-J. Recent progress in carbon quantum dots: Synthesis, properties and applications in photocatalysis. J. Mater. Chem. A 2017, 5, 3717–3734. [Google Scholar] [CrossRef]
- Wang, Y.; Zhu, Y.; Yu, S.; Jiang, C. Fluorescent carbon dots: Rational synthesis, tunable optical properties and analytical applications. RSC Adv. 2017, 7, 40973–40989. [Google Scholar] [CrossRef] [Green Version]
- Dager, A.; Uchida, T.; Maekawa, T.; Tachibana, M. Synthesis and characterization of Mono-disperse Carbon Quantum Dots from Fennel Seeds: Photoluminescence analysis using Machine Learning. Sci. Rep. 2019, 9, 14004. [Google Scholar] [CrossRef]
- Sun, X.; Lei, Y. Fluorescent carbon dots and their sensing applications. TrAC Trends Anal. Chem. 2017, 89, 163–180. [Google Scholar] [CrossRef]
- Hong, G.; Diao, S.; Antaris, A.L.; Dai, H. Carbon Nanomaterials for Biological Imaging and Nanomedicinal Therapy. Chem. Rev. 2015, 115, 10816–10906. [Google Scholar] [CrossRef]
- Feng, T.; Zhao, Y. Preparation of Responsive Carbon Dots for Anticancer Drug Delivery. Methods Mol. Biol. 2019, 2000, 227–234. [Google Scholar]
- Sutanto, H.; Alkian, I.; Romanda, N.; Lewa, I.W.L.; Marhaendrajaya, I.; Triadyaksa, P. High green-emission carbon dots and its optical properties: Microwave power effect. AIP Adv. 2020, 10, 055008. [Google Scholar] [CrossRef]
- Li, X.; Zhang, S.; Kulinich, S.A.; Liu, Y.; Zeng, H. Engineering surface states of carbon dots to achieve controllable luminescence for solid-luminescent composites and sensitive Be2+ detection. Sci. Rep. 2015, 4, 4976. [Google Scholar] [CrossRef]
- Zhou, Y.; Mintz, K.J.; Sharma, S.K.; Leblanc, R.M. Carbon Dots: Diverse Preparation, Application, and Perspective in Surface Chemistry. Langmuir 2019, 35, 9115–9132. [Google Scholar] [CrossRef]
- Ali, H.; Ghosh, S.; Jana, N.R. Fluorescent carbon dots as intracellular imaging probes. WIREs Nanomed. Nanobiotechnol. 2020, 12, e1617. [Google Scholar] [CrossRef]
- Fletcher, A.N. Quinine Sulfate as a Fluorescence Quantum Yield Standard. Photochem. Photobiol. 1969, 9, 439–444. [Google Scholar] [CrossRef] [PubMed]
- Eastman, J.W. Quantitative Spectrofluorimetry-The Fluorescence Quantum Yield of Quinine Sulfate. Photochem. Photobiol. 1967, 6, 55–72. [Google Scholar] [CrossRef]
- Crosby, G.A.; Demas, J.N. Measurement of photoluminescence quantum yields. Review. J. Phys. Chem. 1971, 75, 991–1024. [Google Scholar] [CrossRef]
- Nițică, Ș.; Moldovan, A.I.; Toma, V.; Moldovan, C.S.; Berindan-Neagoe, I.; Știufiuc, G.; Lucaciu, C.M.; Știufiuc, R. PEGylated Gold Nanoparticles with Interesting Plasmonic Properties Synthesized Using an Original, Rapid, and Easy-to-Implement Procedure. J. Nanomater. 2018, 2018, 5954028. [Google Scholar] [CrossRef]
- Știufiuc, G.; Toma, V.; Moldovan, A.; Știufiuc, R. One Pot Microwave Assisted Synthesis of Cyclodextrins Capped Spherical Gold Nanoparticles. Dig. J. Nanomater. Biostruct. 2017, 12, 1089–1095. [Google Scholar]
- Stiufiuc, R.; Iacovita, C.; Nicoara, R.; Stiufiuc, G.; Florea, A.; Achim, M.; Lucaciu, C.M. One-Step Synthesis of PEGylated Gold Nanoparticles with Tunable Surface Charge. J. Nanomater. 2013, 2013, 146031. [Google Scholar] [CrossRef]
- Stiufiuc, R.; Iacovita, C.; Stiufiuc, G.; Florea, A.; Achim, M.; Lucaciu, C.M. A new class of pegylated plasmonic liposomes: Synthesis and characterization. J. Colloid Interface Sci. 2015, 437, 17–23. [Google Scholar] [CrossRef]
- Știufiuc, G.F.; Nițică, Ș.; Toma, V.; Iacoviță, C.; Zahn, D.; Tetean, R.; Burzo, E.; Lucaciu, C.M.; Știufiuc, R.I. Synergistical Use of Electrostatic and Hydrophobic Interactions for the Synthesis of a New Class of Multifunctional Nanohybrids: Plasmonic Magneto-Liposomes. Nanomaterials 2019, 9, 1623. [Google Scholar] [CrossRef] [Green Version]
- Iacovita, C.; Florea, A.; Scorus, L.; Pall, E.; Dudric, R.; Moldovan, A.I.; Stiufiuc, R.; Tetean, R.; Lucaciu, C.M. Hyperthermia, Cytotoxicity, and Cellular Uptake Properties of Manganese and Zinc Ferrite Magnetic Nanoparticles Synthesized by a Polyol-Mediated Process. Nanomaterials 2019, 9, 1489. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carbonaro, C.M.; Corpino, R.; Salis, M.; Mocci, F.; Thakkar, S.V.; Olla, C.; Ricci, P.C. On the Emission Properties of Carbon Dots: Reviewing Data and Discussing Models. C J. Carbon Res. 2019, 5, 60. [Google Scholar] [CrossRef] [Green Version]
- Papavlu, A.P.; Dinca, V.; Filipescu, M.; Dinescu, M. Matrix-Assisted Pulsed Laser Evaporation of Organic Thin Films: Applications in Biology and Chemical Sensors. In Laser Ablation—From Fundamentals to Applications; InTech: Vienna, Austria, 2017. [Google Scholar]
- Ţucureanu, V.; Matei, A.; Avram, A.M. FTIR Spectroscopy for Carbon Family Study. Crit. Rev. Anal. Chem. 2016, 46, 502–520. [Google Scholar] [CrossRef] [PubMed]
- Pelaz, B.; Alexiou, C.; Alvarez-Puebla, R.A.; Alves, F.; Andrews, A.M.; Ashraf, S.; Balogh, L.P.; Ballerini, L.; Bestetti, A.; Brendel, C.; et al. Diverse Applications of Nanomedicine. ACS Nano 2017, 11, 2313–2381. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jung, Y.K.; Shin, E.; Kim, B.-S. Cell Nucleus-Targeting Zwitterionic Carbon Dots. Sci. Rep. 2015, 5, 18807. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pan, L.; Liu, J.; Shi, J. Cancer cell nucleus-targeting nanocomposites for advanced tumor therapeutics. Chem. Soc. Rev. 2018, 47, 6930–6946. [Google Scholar] [CrossRef] [PubMed]
- Belmokhtar, C.A.; Hillion, J.; Ségal-Bendirdjian, E. Staurosporine induces apoptosis through both caspase-dependent and caspase-independent mechanisms. Oncogene 2001, 20, 3354–3362. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Olguín-Albuerne, M.; Domínguez, G.; Morán, J. Effect of Staurosporine in the Morphology and Viability of Cerebellar Astrocytes: Role of Reactive Oxygen Species and NADPH Oxidase. Oxid. Med. Cell. Longev. 2014, 2014, 678371. [Google Scholar] [CrossRef] [Green Version]
- Nishi, K.; Schnier, J.; Brandbury, M. Cell Shape Change Precedes Staurosporine-Induced Stabilization and Accumulation of p27kip1. Exp. Cell Res. 2002, 280, 233–243. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Yang, H.; Liu, H.; Huang, J.; Song, X. Effect of staurosporine on the mobility and invasiveness of lung adenocarcinoma A549 cells: An in vitro study. BMC Cancer 2009, 9, 174. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Montecucco, A.; Zanetta, F.; Biamonti, G. Molecular mechanisms of etoposide. EXCLI J. 2015, 14, 95–108. [Google Scholar] [CrossRef] [PubMed]
- Shin, H.-J.; Kwon, H.-K.; Lee, J.-H.; Anwar, M.A.; Choi, S. Etoposide induced cytotoxicity mediated by ROS and ERK in human kidney proximal tubule cells. Sci. Rep. 2016, 6, 34064. [Google Scholar] [CrossRef] [PubMed]
- Sverchinsky, D.; Nikotina, A.; Komarova, E.; Mikhaylova, E.; Aksenov, N.; Lazarev, V.; Mitkevich, V.; Suezov, R.; Druzhilovskiy, D.; Poroikov, V.; et al. Etoposide-Induced Apoptosis in Cancer Cells Can Be Reinforced by an Uncoupled Link between Hsp70 and Caspase-3. Int. J. Mol. Sci. 2018, 19, 2519. [Google Scholar] [CrossRef] [Green Version]


















| Group | t Test | Wilcoxon-Mann-Whitney | Kolmogorov-Smirnov |
|---|---|---|---|
| Ctrl | 1 | 0.999962061 | 0.063092048 |
| Eto25 | 4.47 × 10−283 | 2.80 × 10−183 | 0.009255334 |
| Eto50 | 0 | 1.58 × 10−193 | 4.92 × 10−5 |
| Stau25 | 0 | 8.51 × 10−225 | 4.18 × 10−5 |
| Stau50 | 0 | 3.49 × 10−220 | 0.003144836 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Moldovan, C.S.; Onaciu, A.; Toma, V.; Marginean, R.; Moldovan, A.; Tigu, A.B.; Stiufiuc, G.F.; Lucaciu, C.M.; Stiufiuc, R.I. Quantifying Cytosolic Cytochrome c Concentration Using Carbon Quantum Dots as a Powerful Method for Apoptosis Detection. Pharmaceutics 2021, 13, 1556. https://0-doi-org.brum.beds.ac.uk/10.3390/pharmaceutics13101556
Moldovan CS, Onaciu A, Toma V, Marginean R, Moldovan A, Tigu AB, Stiufiuc GF, Lucaciu CM, Stiufiuc RI. Quantifying Cytosolic Cytochrome c Concentration Using Carbon Quantum Dots as a Powerful Method for Apoptosis Detection. Pharmaceutics. 2021; 13(10):1556. https://0-doi-org.brum.beds.ac.uk/10.3390/pharmaceutics13101556
Chicago/Turabian StyleMoldovan, Cristian Silviu, Anca Onaciu, Valentin Toma, Radu Marginean, Alin Moldovan, Adrian Bogdan Tigu, Gabriela Fabiola Stiufiuc, Constantin Mihai Lucaciu, and Rares Ionut Stiufiuc. 2021. "Quantifying Cytosolic Cytochrome c Concentration Using Carbon Quantum Dots as a Powerful Method for Apoptosis Detection" Pharmaceutics 13, no. 10: 1556. https://0-doi-org.brum.beds.ac.uk/10.3390/pharmaceutics13101556