Chitosan Oleate Coated PLGA Nanoparticles as siRNA Drug Delivery System
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Methods
2.2.1. Preparation of Chitosan Oleate PLGA Nanoparticles (CS-NPs)
2.2.2. Anti–Tat/Rev siRNAs
2.2.3. siRNAs/CS-NPs Functionalization
2.2.4. Characterization of CS-NPs and SiRNAs/CS-NPs
2.2.5. Qualitative Determination of siRNA-NPs Interaction
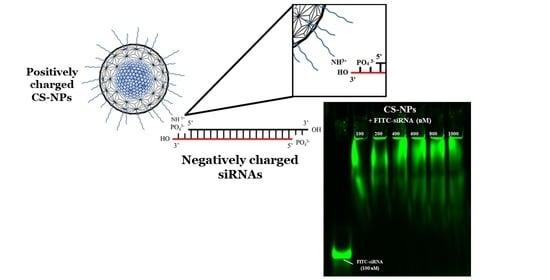
Gel Electrophoresis
Fluorescence Titration Assay
Quantification of Primary Amine Groups
2.2.6. In Vitro Evaluation
- HEPG2 cell line
- PBMCs cell line
Internalization and Flow Cytometric Analyses on HepG2 and PBMCs
- HepG2 cells were washed twice with DPBS 1X, then trypsinized with 0.5 mL/well of Trypsin EDTA 1 × solution in HBSS (Irvine Scientific, Santa Ana, CA, USA), collected in tubes and centrifuged at 160× g for 5 min. Pellets were washed in DPBS 1 X for two times. Finally, cells were fixed with 0.4 mL/tube with IC fixation buffer (Invitrogen, Carlsbad CA, CA, USA) diluted 1:1 in cold DPBS 1× (4 °C). Samples were stored in the fridge, protected from light until FACS (fluorescence activated cell sorting) analysis.
- PBMCs were mechanically detached from the bottom of the well, collected in tubes, centrifuged at 300× g for 5 min, the supernatant discarded, and cells were washed once in cold DPBS 1× (4 °C), followed by another centrifuge step. Then, PBMCs were fixed using the same procedure described above.
Cytotoxicity Test on Human CD14+ Monocytes from Peripheral Blood (hMoCD14+-PB) and Morphological Evaluation
Characterization of Immune Responses of Human PBMCs
3. Results and Discussion
3.1. Characterization of CS-NPs and siRNA/CS-NPs
3.2. Qualitative Determination of siRNA-NPs Interaction
3.2.1. Gel Electrophoresis
3.2.2. Fluorescence Titration Assay
3.2.3. Quantification of Primary Amine Groups
3.3. Internalization and Flow Cytometric Analyses on HepG2 and PBMCs
3.4. Cytotoxicity Test on Human CD14+ Monocytes from Peripheral Blood
3.5. Characterization of Inflammatory Response of Human PBMCs on CS-NPs
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Obbard, D.J.; Gordon, K.H.J.; Buck, A.H.; Jiggins, F.M. The evolution of RNAi as a defence against viruses and transposable elements. Philos. Trans. R. Soc. B 2009, 364, 99–115. [Google Scholar] [CrossRef] [Green Version]
- Elbashir, S.; Harborth, J.; Lendeckel, W.; Yalcin, A.; Weber, K.; Tuschl, T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 2001, 411, 494–498. [Google Scholar] [CrossRef] [PubMed]
- Miele, E.; Spinelli, G.P.; Miele, E.; Di Fabrizio, E.; Ferretti, E.; Tomao, S.; Gulino, A. Nanoparticle-based delivery of small interfering RNA: Challenges for cancer therapy. Int. J. Nanomed. 2012, 7, 3637–3657. [Google Scholar]
- Laganà, A.; Shasha, D.; Croce, C.M. Synthetic RNAs for gene regulation: Design principles and computational tools. Front. Bioeng. Biotechnol. 2014, 2, 65–72. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, B.; Zhong, L.; Weng, Y.; Peng, L.; Huang, Y.; Zhao, Y.; Liang, X.J. Therapeutic siRNA: State of the art. Sig. Transduct. Target Ther. 2020, 5, 101–126. [Google Scholar] [CrossRef]
- Castanotto, D.; Rossi, J.J. The promises and pitfalls of RNA-interference-based therapeutics. Nature 2009, 457, 426–433. [Google Scholar] [CrossRef] [Green Version]
- Pecot, C.V.; Calin, G.A.; Coleman, R.L.; Lopez-Berestein, G.; Sood, A.K. RNA interference in the clinic: Challenges and future directions. Nat. Rev. Cancer 2011, 11, 59–67. [Google Scholar] [CrossRef] [Green Version]
- Kim, D.H.; Rossi, J.J. Strategies for silencing human disease using RNA interference. Nat. Rev. Genet. 2007, 8, 173–184. [Google Scholar] [CrossRef]
- Hu, B.; Weng, Y.; Xia, X.H.; Liang, X.J.; Huang, Y. Clinical advances of siRNA therapeutics. J. Gene Med. 2019, 21, e3097. [Google Scholar] [CrossRef]
- Khan, A.A.; Alanazi, A.M.; Jabeen, M.; Chauhan, A.; Ansari, M.A. Therapeutic potential of functionalized siRNA nanoparticles on regression of liver cancer in experimental mice. Sci. Rep. 2019, 9, 1–16. [Google Scholar] [CrossRef] [Green Version]
- Pai, S.I.; Lin, Y.Y.; Macaes, B.; Meneshian, A.; Hung, C.F.; Wu, T.C. Prospects of RNA interference therapy for cancer. Gene Ther. 2006, 6, 464–477. [Google Scholar] [CrossRef]
- Henney, N.C.; Banach, M.; Penson, P.E. RNA silencing in the management of dyslipidemias. Curr. Atheroscler. Rep. 2021, 23, 1–69. [Google Scholar] [CrossRef]
- Koutsilieri, E.; Rethwilm, A.; Scheller, C. The therapeutic potential of siRNA in gene therapy of neurodegenerative disorders. J. Neural Transm. Suppl. 2007, 72, 43–49. [Google Scholar]
- Burnett, J.C.; Rossi, J.J.; Tiemann, K. Current progress of siRNA/shRNA therapeutics in clinical trials. Biotechnol. J. 2011, 6, 1130–1146. [Google Scholar] [CrossRef] [Green Version]
- Koizumi, M.; Hirota, Y.; Nakayama, M.; Tamura, M.; Obuchi, W.; Kurimoto, A.; Tsuchida, H.; Maeda, H. Design of 2′-O-methyl RNA and DNA double-stranded oligonucleotides: Naturally-occurring nucleotide components with strong RNA interference gene expression inhibitory activity. Nucleosides Nucleotides Nucleic Acids 2020, 39, 292–309. [Google Scholar] [CrossRef] [PubMed]
- Kumar, P.; Parmar, R.G.; Brown, C.R.; Willoughby, J.L.S.; Foster, D.J.; Babu, I.R.; Schofield, S.; Jadhav, V.; Charisse, K.; Nair, J.K.; et al. 5′-Morpholino modification of the sense strand of an siRNA makes it a more effective passenger. Chem. Commun. 2019, 55, 5139–5142. [Google Scholar] [CrossRef] [PubMed]
- Xia, X.; Pollock, N.; Zhou, J.; Rossi, J. Tissue-specific delivery of oligonucleotides. In Oligonucleotide-Based Therapies: Methods and Protocols, Methods in Molecular Biology; Gissberg, O., Zain, R., Lundin, K.E., Eds.; SpringerLink—Humana: New York, NY, USA, 2019; pp. 17–49. [Google Scholar]
- Tao, Y.; Hou, X.; Zuo, F.; Li, X.; Pang, Y.; Jiang, G. Application of nanoparticle-based siRNA and CRISPR/Cas9 delivery systems in gene-targeted therapy. Nanomedicine 2019, 14, 511–514. [Google Scholar] [CrossRef] [Green Version]
- Hu, S.; Li, M.; Akkina, R. Generation of high-titer pseudotyped lentiviral vectors. Methods Mol. Biol. 2019, 1937, 125–134. [Google Scholar] [PubMed]
- Zhou, J.; Rossi, J. Aptamers as targeted therapeutics: Current potential and challenges. Nat. Rev. Drug Discov. 2017, 16, 181–202. [Google Scholar] [CrossRef] [Green Version]
- Setten, R.L.; Rossi, J.J.; Han, S.-P. The current state and future directions of RNAi-based therapeutics. Nat. Rev. Drug Discov. 2019, 18, 421–446. [Google Scholar] [CrossRef]
- Gao, K.; Huang, L. Nonviral methods for siRNA delivery. Mol. Pharm. 2009, 6, 651–658. [Google Scholar] [CrossRef]
- Mohammadinejad, R.; Dehshahri, D.; Madamsetty, V.S.; Zahmatkeshan, M.; Tavakol, S.; Makvandi, P.; Khorsandi, D.; Pardakhty, A.; Ashrafizadeh, M.; Ghasemipour Afshar, E.; et al. In vivo gene delivery mediated by non-viral vectors for cancer therapy. J. Control. Release 2020, 325, 249–275. [Google Scholar] [CrossRef] [PubMed]
- Tortiglione, C.; De la Fuente, J.M. Synthesis of gold nanoparticles for gene silencing. In RNA Interference and Cancer Therapy: Methods and Protocols; Kumar, L.D., Ed.; Methods in Molecular Biology Series; Humana: Totowa, NJ, USA, 2019; pp. 203–214. [Google Scholar]
- He, X.; Yin, F.; Wang, D.; Xiong, L.H.; Kwok, R.T.K.; Gao, P.F.; Zhao, Z.; Lam, J.W.Y.; Yong, K.T.; Li, Z.; et al. AIE featured inorganic-organic Core@Shell nanoparticles for high-efficiency siRNA delivery and real-time monitoring. Nano Lett. 2019, 19, 2272–2279. [Google Scholar] [CrossRef] [PubMed]
- Schroeder, A.; Levins, C.G.; Cortez, C.; Langer, R.; Anderson, D.G. Lipid-based nanotherapeutics for siRNA delivery. J. Intern. Med. 2010, 267, 9–21. [Google Scholar] [CrossRef] [PubMed]
- Zhi, D.; Zhang, S.; Cui, S.; Zhao, Y.; Wang, Y.; Zhao, D. The headgroup evolution of cationic lipids for gene delivery. Bioconjug. Chem. 2013, 24, 487–519. [Google Scholar] [CrossRef] [PubMed]
- Dong, Y.; Siegwart, D.J.; Anderson, D.G. Strategies, design, and chemistry in siRNA delivery systems. Adv. Drug Deliv. Rev. 2019, 144, 133–147. [Google Scholar] [CrossRef] [PubMed]
- Davis, M.E. The first targeted delivery of siRNA in humans via a self-assembling, cyclodextrin polymer-based nanoparticle: From concept to clinic. Mol. Pharm. 2009, 6, 659–668. [Google Scholar] [CrossRef]
- Siegwart, D.J.; Whitehead, K.A.; Nuhn, L.; Sahay, G.; Cheng, H.; Jiang, S.; Ma, M.; Lytton-Jean, A.; Vegas, A.; Fenton, P.; et al. Combinatorial synthesis of chemically diverse core-shell nanoparticles for intracellular delivery. Proc. Natl. Acad. Sci. USA 2011, 108, 12996–13001. [Google Scholar] [CrossRef] [Green Version]
- Karunaratne, D.N.; Jafari, M.; Ranatunga, R.J.; Siriwardhana, A. natural carriers for siRNA delivery. Curr. Pharm. Des. 2015, 21, 4529–4540. [Google Scholar] [CrossRef]
- Singh, B.; Choi, Y.J.; Park, I.K.; Akaike, T.; Cho, C.S. Chemical modification of chitosan with pH-sensitive molecules and specific ligands for efficient DNA transfection and siRNA silencing. J. Nanosci. Nanotechnol. 2014, 14, 564–576. [Google Scholar] [CrossRef]
- Dang, Y.; Guan, J. Nanoparticle-based drug delivery systems for cancer therapy. Smart Mater. Med. 2020, 1, 10–19. [Google Scholar] [CrossRef]
- Serrano-Sevilla, I.; Artiga, Á.; Mitchell, S.G.; De Matteis, L.; De la Fuente, J.M. Natural polysaccharides for siRNA delivery: Nanocarriers based on chitosan, hyaluronic acid, and their derivatives. Molecules 2019, 24, 2570. [Google Scholar] [CrossRef] [Green Version]
- Alameh, M.; Lavertu, M.; Tran-Khanh, N.; Chang, C.Y.; Lesage, F.; Bail, M.; Darras, V.; Chevrier, A.; Buschmann, M.D. siRNA delivery with chitosan: Influence of Chitosan molecular weight, degree of deacetylation, and amine to phosphate ratio on in vitro silencing efficiency, hemocompatibility, biodistribution, and in vivo efficacy. Biomacromolecules 2018, 19, 112–131. [Google Scholar] [CrossRef]
- Hoffmann, N.; Mittnacht, U.; Hartmann, H.; Baumer, Y.; Kjems, J.; Oberhoffner, S.; Schlosshauer, B. Neuronal and glial responses to siRNA-coated nerve guide implants in vitro. Neurosci. Lett. 2011, 494, 14–18. [Google Scholar] [CrossRef]
- Mittnacht, U.; Hartmann, H.; Hein, S.; Oliveira, H.; Dong, M.; Pêgo, A.P.; Kjems, J.; Howard, K.A.; Schlosshauer, B. Chitosan/siRNA nanoparticles biofunctionalize nerve implants and enable neurite outgrowth. Nano Lett. 2010, 10, 3933–3939. [Google Scholar] [CrossRef]
- Cao, Y.; Tan, Y.F.; Wong, Y.S.; Liew, M.; Venkatraman, S. Recent advances in chitosan-based carriers for gene delivery. Mar. Drugs 2019, 17, 381. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Katas, H.; Alpar, O. Development and characterization of chitosan nanoparticles for siRNA delivery. J. Control. Rel. 2006, 115, 2016–2225. [Google Scholar] [CrossRef] [PubMed]
- Patel, P.; Ibrahim, N.M.; Cheng, K. The importance of apparen pKa in the development of nanoparticles encapsulating siRNA and mRNA. Trends Pharmacol. Sci. 2021, 42, 448–459. [Google Scholar] [CrossRef] [PubMed]
- Bonferoni, M.C.; Sandri, G.; Rossi, S.; Usai, D.; Liakos, I.; Garzoni, A.; Fiamma, M.; Zanetti, S.; Athanassiou, A.; Caramella, C.; et al. A novel ionic amphiphilic chitosan derivative as a stabilizer of nanoemulsions: Improvement of antimicrobial activity of Cymbopogon citratus essential oil. Colloids Surf. B Biointerfaces 2017, 152, 385–392. [Google Scholar] [CrossRef]
- Bonferoni, M.C.; Sandri, G.; Dellera, E.; Rossi, S.; Ferrari, F.; Mori, M.; Caramella, C. Ionic polymeric micelles based on chitosan and fatty acids and intended for wound healing. Comparison of linoleic and oleic acid. Eur. J. Pharm. Biopharm. 2014, 87, 101–106. [Google Scholar] [CrossRef]
- Miele, D.; Rossi, S.; Sandri, G.; Vigani, B.; Sorrenti, M.; Giunchedi, P.; Ferrari, F.; Bonferoni, M.C. Chitosan oleate salt as an amphiphilic polymer for the surface modification of Poly-Lactic-Glycolic Acid (PLGA) nanoparticles. Preliminary studies of mucoadhesion and cell interaction properties. Mar Drugs 2018, 16, 447. [Google Scholar] [CrossRef] [Green Version]
- Miele, D.; Catenacci, L.; Sorrenti, M.; Rossi, S.; Sandri, G.; Malavasi, L.; Dacarro, G.; Ferrari, F.; Bonferoni, M.C. Chitosan oleate coated Poly Lactic-Glycolic Acid (PLGA) nanoparticles versus chitosan oleate self-assembled polymeric micelles, Loaded with resveratrol. Mar. Drugs 2019, 17, 515. [Google Scholar] [CrossRef] [Green Version]
- Noel, S.; Liberelle, B.; Robitaille, L.; De Crescenzo, G. Quantification of primary amine groups available for subsequent biofunctionalization of polymer surfaces. Bioconjug. Chem. 2011, 22, 1690–1700. [Google Scholar] [CrossRef] [Green Version]
- Zhou, J.; Lazar, D.; Li, H.; Xia, X.; Satheesan, S.; Charlins, P.; O’Mealy, D.; Akkina, R.; Saayman, S.; Weinberg, M.S.; et al. Receptor-targeted aptamer-siRNA conjugate-directed transcriptional regulation of HIV-1. Theranostics 2018, 8, 1575–1590. [Google Scholar] [CrossRef]
- Zhou, J.; Li, H.; Li, S.; Zaia, J.; Rossi, J.J. Novel dual inhibitory function aptamer-siRNA delivery system for HIV-1 therapy. Mol. Ther. 2008, 16, 1481–1489. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.H.; Behlke, M.A.; Rose, S.D.; Chang, M.S.; Choi, S.; Rossi, J.J. Synthetic dsRNA Dicer substrates enhance RNAi potency and efficacy. Nat. Biotechnol. 2005, 23, 222–228. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rose, S.D.; Kim, D.H.; Amarzguioui, M.; Heidel, J.D.; Collingwood, M.A.; Davis, M.E.; Rossi, J.J.; Behlke, M.A. Functional polarity is introduced by Dicer processing of short substrate RNAs. Nucleic Acids Res. 2005, 33, 4140–4156. [Google Scholar] [CrossRef] [PubMed]
- Instructions: SPDP Crosslinker. Available online: https://www.thermofisher.com/document-connect/document-connect.html?url=https%3A%2F%2Fassets.thermofisher.com%2FTFS-Assets%2FLSG%2Fmanuals%2FMAN0011212_SPDP_CrsLnk_UG.pdf&title=VXNlciBHdWlkZTogIFNQRFAgQ3Jvc3NsaW5rZXJz (accessed on 16 June 2021).
- Lee, D.W.; Yun, K.S.; Ban, H.S.; Choe, W.; Lee, S.K.; Lee, K.Y. Preparation and characterization of chitosan/polyguluronate nanoparticles for siRNA delivery. J. Control Release 2009, 139, 146–152. [Google Scholar] [CrossRef]
- Techaarpornkul, S.; Wongkupasert, S.; Opanasopit, P.; Apirakaramwong, A.; Nunthanid, J.; Ruktanonchai, U. Chitosan-mediated siRNA delivery in vitro: Effect of polymer molecular weight, concentration and salt forms. AAPS PharmSciTech 2011, 11, 64–72. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Strand, S.P.; Danielsen, S.; Christensen, B.; Varum, K.M. Influence of chitosan structure on the formation and stability of DNA-chitosan polyelectrolyte complexes. Biomacromolecules 2005, 6, 3357–3366. [Google Scholar] [CrossRef]
- Layek, B.; Singh, J. 8—Chitosan for DNA and gene therapy. In Chitosan Based Biomaterials; Amber Jennings, J., Bumgardner, J.D., Eds.; Woodhead Publishing: Sawston, UK, 2017; Volume 2, pp. 209–244. [Google Scholar]
- Hoemann, C.D.; Fong, D. 3—Immunological responses to chitosan for biomedical applications. In Chitosan Based Biomaterials; Amber Jennings, J., Bumgardner, J.D., Eds.; Woodhead Publishing: Sawston, UK, 2017; Volume 1, pp. 45–79. [Google Scholar]
- Dellera, E.; Bonferoni, M.C.; Sandri, G.; Rossi, S.; Ferrari, F.; Del Fante, C.; Perotti, C.; Grisoli, P.; Caramella, C. Development of chitosan oleate ionic micelles loaded with silver sulfadiazine to be associated with platelet lysate for application in wound healing. Eur. J. Pharm. Biopharm. 2014, 88, 643–650. [Google Scholar] [CrossRef]
- Tachibana, H.; Kakuta, S.; Yagami, K.; Nagumo, M. Effects of cytokines on the production of nitric oxide in a chondrogenic cell line established from human osteogenic sarcoma. Oral Dis. 2000, 6, 303–309. [Google Scholar] [CrossRef]
- Moran, H.B.T.; Turley, J.L.; Andersson, M.; Lavelle, E.C. Immunomodulatory properties of chitosan polymers. Biomaterials 2018, 184, 1–9. [Google Scholar] [CrossRef]
- Fong, D.; Hoemann, C. Chitosan immunomodulatory properties: Perspectives on the impact of structural properties and dosage. Future Sci. OA 2017, 4, FSO225. [Google Scholar] [CrossRef] [Green Version]
- Dobrovolskaia, M.A.; McNeil, S.E. Immunological properties of engineered nanomaterials. Nat. Nanotechnol. 2007, 2, 469–478. [Google Scholar] [CrossRef]
- Hidalgo, M.A.; Carretta, M.D.; Burgos, R.A. Long chain fatty acids as modulators of immune cells function: Contribution of FFA1 and FFA4 receptors. Front. Physiol. 2021, 12, 668330. [Google Scholar] [CrossRef] [PubMed]
- Santamarina, A.B.; Pisani, L.P.; Baker, E.J.; Marat, A.D.; Valenzuela, C.A.; Miles, E.A.; Calder, P.C. Anti-inflammatory effects of oleic acid and the anthocyanin keracyanin alone and in combination: Effects on monocyte and macrophage responses and the NF-κB pathway. Food Funct. 2021, 12, 7909. [Google Scholar] [CrossRef] [PubMed]
- Nadesh, R.; Narayanan, D.; Sreerekha, P.R.; Vadakumpully, S.; Mony, U.; Koyakkutty, M.; Nair, S.V.; Menon, D. Hematotoxicological analysis of surface-modified and -unmodified chitosan nanoparticles. J. Biomed. Mater. Res. A 2013, 101, 2957–2966. [Google Scholar] [CrossRef]
- Kumar, A.; Vimal, A.; Kumar, A. Why chitosan? From properties to perspective of mucosal drug delivery. Int. J. Biol. Macromol. 2016, 91, 615–622. [Google Scholar] [CrossRef] [PubMed]
- Gavini, E.; Rassu, G.; Ferraro, L.; Generosi, A.; Rau, J.V.; Brunetti, A.; Giunchedi, P.; Dalpiaz, A. Influence of chitosan glutamate on the in vivo intranasal absorption of rokitamycin from microspheres. J. Pharm. Sci. 2011, 100, 1488–1502. [Google Scholar] [CrossRef]
- Yang, S.; Chen, Y.; Ahmadie, R.; Ho, E.A. Advancements in the field of intravaginal SiRNA delivery. J. Controlled Rel. 2013, 167, 29–39. [Google Scholar] [CrossRef] [PubMed]
- Rossi, J.J. RNAi as a treatment for HIV-1 infection. Biotechniques 2006, 40 (Suppl. S4), S25–S29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mobarakeh, V.I.; Modarressi, M.H.; Rahimi, P.; Bolhassani, A.; Arefian, E.; Atyabi, F.; Vahabpour, R. Optimization of chitosan nanoparticles as an anti-HIV siRNA delivery vehicle. Int. J. Biol. Macromol. 2019, 129, 305–315. [Google Scholar] [CrossRef]
- Baxi, K.; Sawarkar, S.; Momin, M.; Patel, V.; Fernandes, T. Vaginal siRNA delivery: Overview on novel delivery approaches. Drug Deliv. Transl. Res. 2020, 10, 962–974. [Google Scholar] [CrossRef] [PubMed]
- Chew, S.A.; Danti, S. Biomaterial-based implantable devices for cancer therapy. Adv. Healthcare Mater. 2017, 6, 2192–2640. [Google Scholar] [CrossRef]
- Perez-Herrero, E.; Fernandez-Medarde, A. Advanced targeted therapies in cancer: Drug nanocarriers, the future of chemotherapy. Eur. J. Pharm. Biopharm. 2015, 93, 52–79. [Google Scholar] [CrossRef] [Green Version]









| Before Dialysis (pH 4.5) | After Dialysis (pH 7.4) | |
|---|---|---|
| PS (nm) | 236.7 ± 4.0 | 333.6 ± 11.5 |
| PI | 0.18 ± 0.03 | 0.14 ± 0.04 |
| Zeta potential, ζ (mV) | 36.49 ± 8.55 | 15.95 ± 3.40 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Miele, D.; Xia, X.; Catenacci, L.; Sorrenti, M.; Rossi, S.; Sandri, G.; Ferrari, F.; Rossi, J.J.; Bonferoni, M.C. Chitosan Oleate Coated PLGA Nanoparticles as siRNA Drug Delivery System. Pharmaceutics 2021, 13, 1716. https://0-doi-org.brum.beds.ac.uk/10.3390/pharmaceutics13101716
Miele D, Xia X, Catenacci L, Sorrenti M, Rossi S, Sandri G, Ferrari F, Rossi JJ, Bonferoni MC. Chitosan Oleate Coated PLGA Nanoparticles as siRNA Drug Delivery System. Pharmaceutics. 2021; 13(10):1716. https://0-doi-org.brum.beds.ac.uk/10.3390/pharmaceutics13101716
Chicago/Turabian StyleMiele, Dalila, Xin Xia, Laura Catenacci, Milena Sorrenti, Silvia Rossi, Giuseppina Sandri, Franca Ferrari, John J. Rossi, and Maria Cristina Bonferoni. 2021. "Chitosan Oleate Coated PLGA Nanoparticles as siRNA Drug Delivery System" Pharmaceutics 13, no. 10: 1716. https://0-doi-org.brum.beds.ac.uk/10.3390/pharmaceutics13101716