DNA-Binding and Cytotoxicity of Copper(I) Complexes Containing Functionalized Dipyridylphenazine Ligands
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Methods and Instrumentation
2.3. Synthesis and Characterizations
General Procedure for the Synthesis of Copper(I) Complexes
2.4. Crystal Structure Determination
2.5. DNA-Binding Studies
2.5.1. Spectroscopic Determination of Binding Affinities
2.5.2. Ethidium Bromide Fluorescence Quenching
2.5.3. Determination of Binding Mode by Viscometry
2.6. Molecular Docking Studies
2.7. Anticancer Studies
3. Results and Discussion
3.1. Synthesis and Characterization
3.2. Crystallographic Studies
3.3. Absorption and Emission Spectra
3.4. DNA-Binding Studies
3.5. Molecular Docking
3.6. Anticancer Studies
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| Dppz | dipyrido[3,2-a:2’,3’-c]phenazine. |
| EtBr | Ethidium bromide. |
| M14 | melanoma skin cancer cell line. |
| MCF-7 | breast cancer cell line. |
References
- Wong, E.; Giandomenico, C.M. Current Status of Platinum-Based Antitumor Drugs. Chem. Rev. 1999, 99, 2451–2466. [Google Scholar] [CrossRef] [PubMed]
- Abu-Surrah, A.S.; Kettunen, M. Platinum Group Antitumor Chemistry: Design and development of New Anticancer Drugs Complementary to Cisplatin. Curr. Med. Chem. 2006, 13, 1337–1357. [Google Scholar] [CrossRef] [PubMed]
- Rabik, C.A.; Dolan, M.E. Molecular mechanisms of resistance and toxicity associated with platinating agents. Cancer Treat. Rev. 2007, 33, 9–23. [Google Scholar] [CrossRef] [Green Version]
- Heffeter, P.; Jungwirth, U.; Jakupec, M.; Hartinger, C.; Galanski, M.; Elbling, L.; Micksche, M.; Keppler, B.; Berger, W. Resistance against novel anticancer metal compounds: Differences and similarities. Drug Resist. Updates 2008, 11, 1–16. [Google Scholar] [CrossRef]
- Yu, G.; Yu, S.; Saha, M.L.; Zhou, J.; Cook, T.R.; Yung, B.C.; Chen, J.; Mao, Z.; Zhang, F.; Zhou, Z.; et al. A discrete organoplatinum(II) metallacage as a multimodality theranostic platform for cancer photochemotherapy. Nat. Commun. 2018, 9, 4335. [Google Scholar] [CrossRef] [PubMed]
- Trudu, F.; Amato, F.; Vanhara, P.; Pivetta, T.; Pena-Mendez, E.M.; Havel, J. Coordination compounds in cancer: Past, present and perspectives. J. Appl. Biomed. 2015, 13, 79–103. [Google Scholar] [CrossRef]
- Mookerjee, A.; Basu, J.M.; Majumder, S.; Chatterjee, S.; Panda, G.S.; Dutta, P.; Pal, S.; Mukherjee, P.; Efferth, T.; Roy, S.; et al. A novel copper complex induces ROS generation in doxorubicin resistant Ehrlich ascitis carcinoma cells and increases activity of antioxidant enzymes in vital organs in vivo. BMC Cancer 2006, 6, 267. [Google Scholar] [CrossRef] [Green Version]
- Marzano, C.; Pellei, M.; Tisato, F.; Santini, C. Copper Complexes as Anticancer Agents. Anticancer Agents Med. Chem. 2009, 9, 185–211. [Google Scholar] [CrossRef] [PubMed]
- Gubendran, A.; Rajesh, J.; Anitha, K.; Athappan, P. Synthesis, characterization, DNA-binding and cleavage studies of polypyridyl copper(II) complexes. J. Mol. Struct. 2014, 1075, 419–429. [Google Scholar] [CrossRef]
- Amali, I.B.; Kesavan, M.P.; Vijayakumar, V.; Gandhi, N.I.; Ragesh, J.; Rajagopal, G. Structural analysis, antimicrobial and cytotoxic studies on new metal(II) complexes containing N2O2 donor Schiff base ligand. J. Mol. Struct. 2019, 1183, 342–350. [Google Scholar] [CrossRef]
- Corona-Motolinia, N.D.; Martínez-Valencia, B.; Noriega, L.; Sánchez-Gaytán, B.L.; Méndez-Rojas, M.A.; Melendez, F.J.; Castro, M.E.; González-Vergara, E. Synthesis, Crystal Structure, and Computational Methods of Vanadium and Copper Compounds as Potential Drugs for Cancer Treatment. Molecules 2020, 25, 4679. [Google Scholar] [CrossRef]
- Duncan, C.; White, A.R. Copper complexes as therapeutic agents. Metallomics 2012, 4, 127–138. [Google Scholar] [CrossRef]
- Marzano, C.; Gandin, V.; Pellei, M.; Colavito, D.; Papini, G.; Lobbia, G.G.; del Giudice, E.; Porchia, M.; Tisato, F.; Santini, C. In Vitro Antitumor Activity of the Water Soluble Copper(I) Complexes Bearing the Tris(hydroxymethyl)phosphine Ligand. J. Med. Chem. 2008, 51, 798–808. [Google Scholar] [CrossRef]
- Gandin, V.; Pellei, M.; Tisato, F.; Porchia, M.; Santini, C.; Marzano, C. A novel copper complex induces paraptosis in colon cancer cells via the activation of ER stress signalling. J. Cell Mol. Med. 2012, 16, 142–151. [Google Scholar] [CrossRef] [Green Version]
- Zanella, A.; Gandin, V.; Porchia, M.; Refosco, F.; Tisato, F.; Sorrentino, F.; Scutari, G.; Rigobello, M.P.; Marzano, C. Cytotoxicity in human cancer cells and mitochondrial dysfunction induced by a series of new copper(I) complexes containing tris(2-cyanoethyl)phosphines. Investig. New Drugs 2011, 29, 1213–1223. [Google Scholar] [CrossRef]
- Starosta, R.; Stokowa, K.; Florek, M.; Król, J.; Chwiłkowska, A.; Kulbacka, J.; Saczko, J.; Skała, J.; Jeżowska-Bojczuk, M. Free Radicals and Antioxidant Defense in Antitumor Therapy, in Antioxidant Enzyme. J. Inorg. Biochem. 2011, 105, 1102–1108. [Google Scholar] [CrossRef]
- Komarnicka, U.K.; Starosta, R.; Płotek, M.; de Almeida, R.F.M.; Jeżowska-Bojczuk, M.; Kyzioł, A. Copper(i) complexes with phosphine derived from sparfloxacin. Part II: A first insight into the cytotoxic action mode. Dalton Trans. 2016, 45, 5052–5063. [Google Scholar] [CrossRef] [PubMed]
- Kyzioł, A.; Cierniak, A.; Gubernator, J.; Markowski, A.; Jeżowska-Bojczuk, M.; Komarnicka, U.K. Copper(i) complexes with phosphine derived from sparfloxacin. Part III: Multifaceted cell death and preliminary study of liposomal formulation of selected copper(i) complexes. Dalton Trans. 2018, 47, 1981–1992. [Google Scholar] [CrossRef] [PubMed]
- Mashat, K.; Babgi, B.; Hussien, M.; Arshad, M.; Abdellattif, M. Synthesis, structures, DNA-binding and anticancer activities of some copper(I)-phosphine complexes. Polyhedron 2019, 158, 164–172. [Google Scholar] [CrossRef]
- Babgi, B.A.; Mashat, K.H.; Abdellattif, M.H.; Arshad, M.N.; Alzahrani, K.A.; Asiri, A.M.; Humphrey, M.G.; Hussien, M. Synthesis, structures, DNA-binding, cytotoxicity and molecular docking of CuBr(PPh3)(diimine). Polyhedron 2020, 192, 114847. [Google Scholar] [CrossRef]
- Gujadhur, R.; Venkataraman, D.; Kintigh, J.T. Formation of aryl-nitrogen bonds using a soluble copper(I) catalyst. Tetrahedron Lett. 2001, 42, 4791–4793. [Google Scholar] [CrossRef]
- Dickeson, J.E.; Summers, L.A. Derivatives of 1,10-Phenanthroline-5,6-quinone. Aust. J. Chem. 1970, 23, 1023–1027. [Google Scholar] [CrossRef]
- Arancibia, A.; Concepcion, J.; Daire, N.; Leiva, G.; Leiva, A.; Loeb, B.; Río, R.; Díaz, R.; Francois, A.; Saldivia, M. Electronic effects of donors and acceptor substituents on dipyrido(3,2-:2′,3′-c)phenazine (dppz). J. Coord. Chem. 2001, 54, 323–336. [Google Scholar] [CrossRef] [Green Version]
- Zúñiga, C.; Crivelli, I.; Loeb, B. Synthesis, characterization, spectroscopic and electrochemical studies of donor–acceptor ruthenium(II) polypyridine ligand derivatives with potential NLO applications. Polyhedron 2015, 85, 511–518. [Google Scholar] [CrossRef]
- Kumar, K.A.; Reddy, K.L.; Vidhisha, S.; Satyanarayana, S. Synthesis, characterization and DNA binding and photocleavage studies of [Ru(bpy)2BDPPZ]2+, [Ru(dmb)2BDPPZ]2+ and [Ru(phen)2BDPPZ]2+ complexes and their antimicrobial activity. Appl. Organomet. Chem. 2009, 23, 409–420. [Google Scholar] [CrossRef]
- Kleineweischede, A.; Mattay, J. Synthesis of Amino- and Bis(bromomethyl)-Substitued Bi- and TetradentateN-Heteroaromatic Ligands: Building Blocks for Pyrazino-Functionalized Fullerene Dyads. Eur. J. Org. Chem. 2006, 2006, 947–957. [Google Scholar] [CrossRef]
- Al-Masri, H.T.; Emwas, A.M.; Al-Talla, Z.A.; Alkordi, M.H. Synthesis and Characterization of New N-(Diphenylphosphino)-Naphthylamine Chalcogenides: X-Ray Structures of (1-NHC10H7)P(Se)Ph2 and Ph2P(S)OP(S)Ph2. Phosphorus Sulfur Silicon Relat. Elem. 2012, 187, 1082–1090. [Google Scholar]
- CrysAlis PRO; Agilent Technologies: Yarnton, UK, 2012.
- Sheldrick, G.M. Crystal structure refinement with SHELXL. Acta Crystallogr. 2015, C71, 3–8. [Google Scholar]
- Farrugia, L.J. WinGXsuite for small-molecule single-crystal crystallography. J. Appl. Crystallogr. 1999, 32, 837–838. [Google Scholar] [CrossRef]
- Spek, A.L. Single-crystal structure validation with the program PLATON. J. Appl. Crystallogr. 2003, 36, 7–13. [Google Scholar] [CrossRef] [Green Version]
- Farrugia, L.J. WinGX and ORTEP for Windows: An update. J. Appl. Crystallogr. 2012, 45, 849–854. [Google Scholar] [CrossRef]
- Devi, C.S.; Thulasiram, B.; Satyanarayana, S.; Nagababu, P. Analytical Techniques Used to Detect DNA Binding Modes of Ruthenium(II) Complexes with Extended Phenanthroline Ring. J. Fluoresc. 2017, 27, 2119–2130. [Google Scholar] [CrossRef]
- Villarreal, W.; Colina-Vegas, L.; Visbal, G.; Corona, O.; Corrêa, R.S.; Ellena, J.; Cominetti, M.R.; Batista, A.A.; Navarro, M. Copper(I)–Phosphine Polypyridyl Complexes: Synthesis, Characterization, DNA/HSA Binding Study, and Antiproliferative Activity. Inorg. Chem. 2017, 56, 3781–3793. [Google Scholar] [CrossRef]
- Muanza, D.N.; Kim, B.W.; Euler, K.L.; Williams, L. Antibacterial and Antifungal Activities of Nine Medicinal Plants from Zaire. Int. J. Pharmacog. 1994, 32, 337–345. [Google Scholar] [CrossRef]
- Pezzuto, J.M.; Che, C.-T.; McPherson, D.D.; Zhu, J.-P.; Topcu, G.; Erdelmeier, C.A.J.; Cordell, G.A. DNA as an Affinity Probe Useful in the Detection and Isolation of Biologically Active Natural Products. J. Natur. Prod. 1991, 54, 1522–1530. [Google Scholar] [CrossRef] [PubMed]
- Skehan, P.; Storeng, R.; Scudiero, D.; Monks, A.; McMahon, J.; Vistica, D.; Warren, J.; Bokesch, H.; Kenney, S.; Boyd, M. New Colorimetric Cytotoxicity Assay for Anticancer-Drug Screening. J. Natl. Cancer Inst. 1990, 82, 1107–1112. [Google Scholar] [CrossRef] [PubMed]
- Spek, A.A. PLATONSQUEEZE: A tool for the calculation of the disordered solvent contribution to the calculated structure factors. Acta Crystallogr. 2015, C71, 9–18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Musina, E.I.; Shamsieva, A.V.; Strelnik, I.D.; Gerasimova, T.P.; Krivolapov, D.B.; Kolesnikov, I.E.; Grachova, E.V.; Tunik, S.P.; Bannwarth, C.; Grimme, S.; et al. Synthesis of novel pyridyl containing phospholanes and their polynuclear luminescent copper(i) complexes. Dalton Trans. 2016, 45, 2250–2260. [Google Scholar] [CrossRef]
- Czerwieniec, R.; Kowalski, K.; Yersin, H. Highly efficient thermally activated fluorescence of a new rigid Cu(I) complex [Cu(dmp)(phanephos)]+. Dalton Trans. 2013, 42, 9826–9830. [Google Scholar] [CrossRef] [Green Version]
- Kalantari, R.; Asadi, Z. DNA/BSA binding of a new oxovanadium (IV) complex of glycylglycine derivative Schiff base ligand. J. Mol. Struct. 2020, 1219, 128664. [Google Scholar] [CrossRef]
- Seferoğlu, Z.; Mahmoud, M.M.A.; Ihmels, H. Studies of the binding interactions of dicationic styrylimidazo[1,2-a]pyridinium dyes with duplex and quadruplex DNA. Dyes Pigments 2016, 125, 241–248. [Google Scholar] [CrossRef]
- Selvakumar, B.; Rajendiran, V.; Maheswari, P.U.; Stoeckli-Evans, H.; Palaniandavar, M. Structures, spectra, and DNA-binding properties of mixed ligand copper(II) complexes of iminodiacetic acid: The novel role of diimine co-ligands on DNA conformation and hydrolytic and oxidative double strand DNA cleavage. J. Inorg. Biochem. 2006, 100, 316–330. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Busto, N.; Martínez-Alonso, M.; Leal, J.M.; Rodríguez, A.M.; Domínguez, F.; Acuña, M.I.; Espino, G.; García, B. Monomer–Dimer Divergent Behavior toward DNA in a Half-Sandwich Ruthenium(II) Aqua Complex. Antiproliferative Biphasic Activity. Organometallics 2015, 34, 319–327. [Google Scholar] [CrossRef]
- Santini, C.; Pellei, M.; Gandin, V.; Porchia, M.; Tisato, F.; Marzano, C. Advances in Copper Complexes as Anticancer Agents. Chem. Rev. 2014, 114, 815–862. [Google Scholar] [CrossRef] [PubMed]










| Complex | Absorbance | Emission | ||
|---|---|---|---|---|
| λmax (nm) | ϵ (103 M−1 cm−1) | λem, (nm) | Relative Intensity | |
| Cu-1 | 273 359 | 47.69 8.61 | 416 | 0.38 |
| Cu-2 | 289 364 | 52.67 13.89 | 436 | 0.66 |
| Cu-3 | 277 368 | 65.36 13.39 | 437 | 0.50 |
| Cu-4 | 283 370 387 | 64.98 14.86 13.85 | 437 | 0.64 |
| Cu-5 | 281 369 388 | 35.82 8.55 9.68 | 418 | 1.00 |
| Complex | Substituent | UV-vis Spectroscopic Titration | Fluorescence Quenching Assay | ||
|---|---|---|---|---|---|
| Kb | Scaled Kb | Ksv | Scaled Ksv | ||
| Cu-1 | H | 4.00 × 106 | 20 | 7.73 × 103 | 1.00 |
| Cu-2 | NO2 | 4.44 × 106 | 22 | 8.93 × 103 | 1.16 |
| Cu-3 | CN | 8.00 × 105 | 4 | 1.00 × 104 | 1.29 |
| Cu-4 | Benzoyl | 8.00 × 105 | 4 | 5.53 × 103 | 0.72 |
| Cu-5 | Dimethyl | 2.00 × 105 | 1 | 6.36 × 103 | 0.82 |
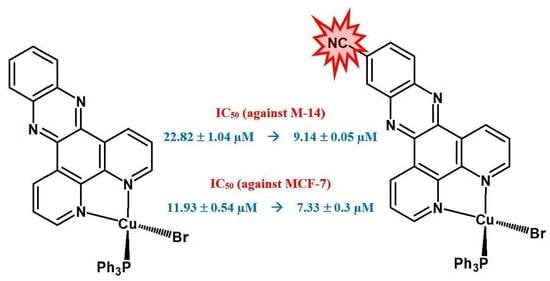
| Compounds | IC50 (µM) | |
|---|---|---|
| M-14 | MCF-7 | |
| cisplatin | 6.29 ± 0.05 | 8.69 ± 0.43 |
| Dppz-NO2 | 57.64 ± 0.05 | - |
| Dppz-CN | 45.20 ± 0.05 | - |
| Cu-1 | 22.82 ± 1.04 | 11.93 ± 0.54 |
| Cu-2 | 13.83 ± 0.46 | 9.85 ± 0.10 |
| Cu-3 | 9.14 ± 0.05 | 7.33 ± 0.38 |
| Cu-4 | 12.88 ± 0.09 | 8.43 ± 0.62 |
| Cu-5 | 14.76 ± 0.31 | 11.52 ± 0.52 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alsaedi, S.; Babgi, B.A.; Abdellattif, M.H.; Arshad, M.N.; Emwas, A.-H.M.; Jaremko, M.; Humphrey, M.G.; Asiri, A.M.; Hussien, M.A. DNA-Binding and Cytotoxicity of Copper(I) Complexes Containing Functionalized Dipyridylphenazine Ligands. Pharmaceutics 2021, 13, 764. https://0-doi-org.brum.beds.ac.uk/10.3390/pharmaceutics13050764
Alsaedi S, Babgi BA, Abdellattif MH, Arshad MN, Emwas A-HM, Jaremko M, Humphrey MG, Asiri AM, Hussien MA. DNA-Binding and Cytotoxicity of Copper(I) Complexes Containing Functionalized Dipyridylphenazine Ligands. Pharmaceutics. 2021; 13(5):764. https://0-doi-org.brum.beds.ac.uk/10.3390/pharmaceutics13050764
Chicago/Turabian StyleAlsaedi, Sammar, Bandar A. Babgi, Magda H. Abdellattif, Muhammad N. Arshad, Abdul-Hamid M. Emwas, Mariusz Jaremko, Mark G. Humphrey, Abdullah M. Asiri, and Mostafa A. Hussien. 2021. "DNA-Binding and Cytotoxicity of Copper(I) Complexes Containing Functionalized Dipyridylphenazine Ligands" Pharmaceutics 13, no. 5: 764. https://0-doi-org.brum.beds.ac.uk/10.3390/pharmaceutics13050764