Ectopic Odorant Receptor Responding to Flavor Compounds: Versatile Roles in Health and Disease
Abstract
:1. Introduction
2. Main Olfactory System
3. ORs Are Not Only Expressed in OE
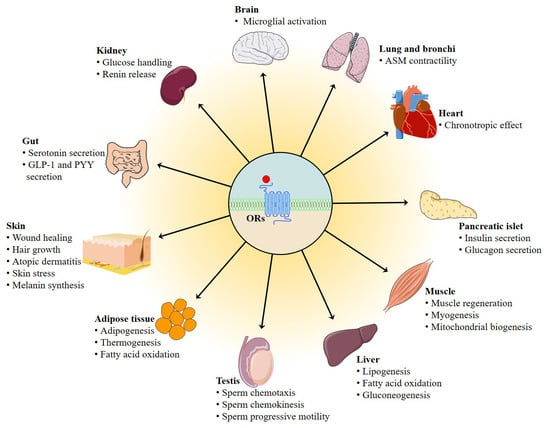
4. Biological Functions of Ectopic ORs Responding to Odorant Compounds in the Non-Chemosensory Tissues
4.1. Role of Ectopic OR in the Testis
4.2. Role of Ectopic ORs in the Muscle
4.3. Role of Ectopic OR in the Adipose Tissue
4.4. Role of Ectopic ORs in the Liver
4.5. Role of Ectopic ORs in the Gut
4.6. Role of Ectopic ORs in the Kidney
4.7. Role of Ectopic ORs in the Skin
4.8. Role of Ectopic ORs in the Lung and Bronchi
4.9. Role of Ectopic ORs in the Cardiovascular System
4.10. Role of Ectopic ORs in the Pancreatic Islet
4.11. Role of Ectopic ORs in the Brain
4.12. Role of Ectopic ORs in the Cancer Tissue
4.13. Role of Ectopic ORs in the Coronavirus Disease 2019
5. Ligands of Ectopic ORs
6. Conclusions and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zozulya, S.; Echeverri, F.; Nguyen, T. The human olfactory receptor repertoire. Genome Biol. 2001, 2, research0018.1. [Google Scholar] [CrossRef]
- Zhang, X.; Firestein, S. The olfactory receptor gene superfamily of the mouse. Nat. Neurosci. 2002, 5, 124–133. [Google Scholar] [CrossRef] [PubMed]
- Buck, L.; Axel, R. A novel multigene family may encode odorant receptors: A molecular basis for odor recognition. Cell 1991, 65, 175–187. [Google Scholar] [CrossRef]
- Kaupp, U.B. Olfactory signalling in vertebrates and insects: Differences and commonalities. Nat. Rev. Neurosci. 2010, 11, 188–200. [Google Scholar] [CrossRef] [PubMed]
- Spehr, M.; Gisselmann, G.; Poplawski, A.; Riffell, J.A.; Wetzel, C.H.; Zimmer, R.K.; Hatt, H. Identification of a testicular odorant receptor mediating human sperm chemotaxis. Science 2003, 299, 2054–2058. [Google Scholar] [CrossRef]
- Fleischer, J.; Bumbalo, R.; Bautze, V.; Strotmann, J.; Breer, H. Expression of odorant receptor Olfr78 in enteroendocrine cells of the colon. Cell Tissue Res. 2015, 361, 697–710. [Google Scholar] [CrossRef]
- Pluznick, J.L.; Protzko, R.J.; Gevorgyan, H.; Peterlin, Z.; Sipos, A.; Han, J.; Brunet, I.; Wan, L.X.; Rey, F.; Wang, T.; et al. Olfactory receptor responding to gut microbiota-derived signals plays a role in renin secretion and blood pressure regulation. Proc. Natl. Acad. Sci. USA 2013, 110, 4410–4415. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Drutel, G.; Arrang, J.M.; Diaz, J.; Wisnewsky, C.; Schwartz, K.; Schwartz, J.C. Cloning of ol1, a putative olfactory receptor and its expression in the developing rat-heart. Recept. Channels 1995, 3, 33–40. [Google Scholar]
- Griffin, C.A.; Kafadar, K.A.; Pavlath, G.K. MOR23 promotes muscle regeneration and regulates cell adhesion and migration. Dev. Cell 2009, 17, 649–661. [Google Scholar] [CrossRef] [Green Version]
- Kang, N.; Bahk, Y.Y.; Lee, N.; Jae, Y.; Cho, Y.H.; Ku, C.R.; Byun, Y.; Lee, E.J.; Kim, M.S.; Koo, J. Olfactory receptor Olfr544 responding to azelaic acid regulates glucagon secretion in alpha-cells of mouse pancreatic islets. Biochem. Biophys. Res. Commun. 2015, 460, 616–621. [Google Scholar] [CrossRef]
- Gu, X.; Karp, P.H.; Brody, S.L.; Pierce, R.A.; Welsh, M.J.; Holtzman, M.J.; Ben-Shahar, Y. Chemosensory functions for pulmonary neuroendocrine cells. Am. J. Respir. Cell Mol. 2014, 50, 637–646. [Google Scholar] [CrossRef] [Green Version]
- Wu, C.; Jia, Y.; Lee, J.H.; Kim, Y.; Sekharan, S.; Batista, V.S.; Lee, S.J. Activation of OR1A1 suppresses PPAR-gamma expression by inducing HES-1 in cultured hepatocytes. Int. J. Biochem. Cell Biol. 2015, 64, 75–80. [Google Scholar] [CrossRef]
- Otaki, J.M.; Yamamoto, H.; Firestein, S. Odorant receptor expression in the mouse cerebral cortex. J. Neurobiol. 2004, 58, 315–327. [Google Scholar] [CrossRef]
- Tham, E.H.; Dyjack, N.; Kim, B.E.; Rios, C.; Seibold, M.A.; Leung, D.Y.M.; Goleva, E. Expression and function of the ectopic olfactory receptor OR10G7 in patients with atopic dermatitis. J. Allergy Clin. Immunol. 2019, 143, 1838–1848 e1834. [Google Scholar] [CrossRef]
- Malki, A.; Fiedler, J.; Fricke, K.; Ballweg, I.; Pfaffl, M.W.; Krautwurst, D. Class I odorant receptors, TAS1R and TAS2R taste receptors, are markers for subpopulations of circulating leukocytes. J. Leukoc. Biol. 2015, 97, 533–545. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Geithe, C.; Andersen, G.; Malki, A.; Krautwurst, D. A butter aroma recombinate activates human class-I odorant receptors. J. Agric. Food Chem. 2015, 63, 9410–9420. [Google Scholar] [CrossRef]
- Marcinek, P.; Geithe, C.; Krautwurst, D. Chemosensory G Protein-Coupled Receptors (GPCR) in Blood Leukocytes. In Taste and Smell; Krautwurst, D., Ed.; Springer International Publishing: Cham, Switzerland, 2017; pp. 151–173. [Google Scholar]
- Tong, T.; Park, J.; Moon, C.; Park, T. Regulation of adipogenesis and thermogenesis through mouse olfactory receptor 23 stimulated by alpha-cedrene in 3T3-L1 cells. Nutrients 2018, 10, 1781. [Google Scholar] [CrossRef] [Green Version]
- Neuhaus, E.M.; Zhang, W.; Gelis, L.; Deng, Y.; Noldus, J.; Hatt, H. Activation of an olfactory receptor inhibits proliferation of prostate cancer cells. J. Biol. Chem. 2009, 284, 16218–16225. [Google Scholar] [CrossRef] [Green Version]
- Thach, T.T.; Wu, C.; Hwang, K.Y.; Lee, S.J. Azelaic acid induces mitochondrial biogenesis in skeletal muscle by activation of olfactory receptor 544. Front. Physiol. 2020, 11, 329. [Google Scholar] [CrossRef] [Green Version]
- Malnic, B.; Hirono, J.; Sato, T.; Buck, L.B. Combinatorial receptor codes for odors. Cell 1999, 96, 713–723. [Google Scholar] [CrossRef] [Green Version]
- Nara, K.; Saraiva, L.R.; Ye, X.; Buck, L.B. A large-scale analysis of odor coding in the olfactory epithelium. J. Neurosci. 2011, 31, 9179–9191. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ronnett, G.V.; Moon, C. G proteins and olfactory signal transduction. Annu. Rev. Physiol. 2002, 64, 189–222. [Google Scholar] [CrossRef] [PubMed]
- Parmentier, M.; Libert, F.; Schurmans, S.; Schiffmann, S.; Lefort, A.; Eggerickx, D.; Ledent, C.; Mollereau, C.; Gerard, C.; Perret, J.; et al. Expression of members of the putative olfactory receptor gene family in mammalian germ cells. Nature 1992, 355, 453–455. [Google Scholar] [CrossRef] [PubMed]
- Vanderhaeghen, P.; Schurmans, S.; Vassart, G.; Parmentier, M. Olfactory receptors are displayed on dog mature sperm cells. J. Cell Biol. 1993, 123, 1441–1452. [Google Scholar] [CrossRef]
- Vanderhaeghen, P.; Schurmans, S.; Vassart, G.; Parmentier, M. Specific repertoire of olfactory receptor genes in the male germ cells of several mammalian species. Genomics 1997, 39, 239–246. [Google Scholar] [CrossRef]
- Kang, N.; Koo, J. Olfactory receptors in non-chemosensory tissues. BMB Rep. 2012, 45, 612–622. [Google Scholar] [CrossRef] [Green Version]
- Kerslake, R.; Hall, M.; Randeva, H.S.; Spandidos, D.A.; Chatha, K.; Kyrou, I.; Karteris, E. Coexpression of peripheral olfactory receptors with SARSCoV2 infection mediators: Potential implications beyond loss of smell as a COVID19 symptom. Int. J. Mol. Med. 2020, 46, 949–956. [Google Scholar] [CrossRef]
- Choi, Y.; Hur, C.G.; Park, T. Induction of olfaction and cancer-related genes in mice fed a high-fat diet as assessed through the mode-of-action by network identification analysis. PLoS ONE 2013, 8, e56610. [Google Scholar] [CrossRef] [Green Version]
- Flegel, C.; Vogel, F.; Hofreuter, A.; Schreiner, B.S.P.; Osthold, S.; Veitinger, S.; Becker, C.; Brockmeyer, N.H.; Muschol, M.; Wennemuth, G.; et al. Characterization of the olfactory receptors expressed in human spermatozoa. Front. Mol. Biosci. 2016, 2, 73. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pluznick, J.L.; Zou, D.J.; Zhang, X.; Yan, Q.; Rodriguez-Gil, D.J.; Eisner, C.; Wells, E.; Greer, C.A.; Wang, T.; Firestein, S.; et al. Functional expression of the olfactory signaling system in the kidney. Proc. Natl. Acad. Sci. USA 2009, 106, 2059–2064. [Google Scholar] [CrossRef] [Green Version]
- Kalbe, B.; Schlimm, M.; Wojcik, S.; Philippou, S.; Massberg, D.; Jansen, F.; Scholz, P.; Luebbert, H.; Ubrig, B.; Osterloh, S.; et al. Olfactory signaling components and olfactory receptors are expressed in tubule cells of the human kidney. Arch. Biochem. Biophys. 2016, 610, 8–15. [Google Scholar] [CrossRef]
- Foster, S.R.; Roura, E.; Thomas, W.G. Extrasensory perception: Odorant and taste receptors beyond the nose and mouth. Pharmacol. Ther. 2014, 142, 41–61. [Google Scholar] [CrossRef]
- Maßberg, D.; Hatt, H. Human olfactory receptors: Novel cellular functions outside of the nose. Physiol. Rev. 2018, 98, 1739–1763. [Google Scholar] [CrossRef]
- Wu, C.; Hwang, S.H.; Jia, Y.; Choi, J.; Kim, Y.J.; Choi, D.; Pathiraja, D.; Choi, I.G.; Koo, S.H.; Lee, S.J. Olfactory receptor 544 reduces adiposity by steering fuel preference toward fats. J. Clin. Investig. 2017, 127, 4118–4123. [Google Scholar] [CrossRef] [Green Version]
- Tong, T.; Kim, M.; Park, T. Alpha-Cedrene, a newly identified ligand of mor23, increases skeletal muscle mass and strength. Mol. Nutr. Food Res. 2018, 62, e1800173. [Google Scholar] [CrossRef]
- Chen, Z.; Zhao, H.; Fu, N.; Chen, L. The diversified function and potential therapy of ectopic olfactory receptors in non-olfactory tissues. J. Cell Physiol. 2018, 233, 2104–2115. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.; Thach, T.T.; Kim, Y.J.; Lee, S.J. Olfactory receptor 43 reduces hepatic lipid accumulation and adiposity in mice. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2019, 1864, 489–499. [Google Scholar] [CrossRef]
- Zhang, X.M.; Rogers, M.; Tian, H.K.; Zhang, X.H.; Zou, D.J.; Jian, L.; Ma, M.H.; Shepherd, G.M.; Firestein, S.J. High-throughput microarray detection of olfactory receptor gene expression in the mouse. Proc. Natl. Acad. Sci. USA 2004, 101, 14168–14173. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weyand, I.; Godde, M.; Frings, S.; Weiner, J.; Muller, F.; Altenhofen, W.; Hatt, H.; Kaupp, U.B. Cloning and functional expression of a cyclic-nucleotide-gated channel from mammalian sperm. Nature 1994, 368, 859–863. [Google Scholar] [CrossRef]
- Gautier-Courteille, C.; Salanova, M.; Conti, M. The olfactory adenylyl cyclase III is expressed in rat germ cells during spermiogenesis. Endocrinology 1998, 139, 2588–2599. [Google Scholar] [CrossRef] [PubMed]
- Veitinger, T.; Riffell, J.R.; Veitinger, S.; Nascimento, J.M.; Triller, A.; Chandsawangbhuwana, C.; Schwane, K.; Geerts, A.; Wunder, F.; Berns, M.W.; et al. Chemosensory Ca2+ dynamics correlate with diverse behavioral phenotypes in human sperm. J. Biol. Chem. 2011, 286, 17311–17325. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fukuda, N.; Yomogida, K.; Okabe, M.; Touhara, K. Functional characterization of a mouse testicular olfactory receptor and its role in chemosensing and in regulation of sperm motility. J. Cell Sci. 2004, 117, 5835–5845. [Google Scholar] [CrossRef] [Green Version]
- Khakimov, B.; Jespersen, B.M.; Engelsen, S.B. Comprehensive and comparative metabolomic profiling of wheat, barley, oat and rye using gas chromatography-mass spectrometry and advanced chemometrics. Foods 2014, 3, 569–585. [Google Scholar] [CrossRef] [Green Version]
- Kalbe, B.; Osterloh, M.; Schulz, V.M.; Altmuller, J.; Becker, C.; Osterloh, S.; Hatt, H. OR2H2 regulates the differentiation of human myoblast cells by its ligand aldehyde 13–13. Arch. Biochem. Biophys. 2018, 645, 72–80. [Google Scholar] [CrossRef] [PubMed]
- Tong, T.; Ryu, S.E.; Min, Y.; de March, C.A.; Bushdid, C.; Golebiowski, J.; Moon, C.; Park, T. Olfactory receptor 10J5 responding to alpha-cedrene regulates hepatic steatosis via the cAMP-PKA pathway. Sci. Rep. 2017, 7, 9471. [Google Scholar] [CrossRef] [Green Version]
- Li, E.; Shan, H.; Chen, L.; Long, A.; Zhang, Y.; Liu, Y.; Jia, L.; Wei, F.; Han, J.; Li, T.; et al. OLFR734 mediates glucose metabolism as a receptor of asprosin. Cell Metab. 2019, 30, 319–328 e318. [Google Scholar] [CrossRef]
- Head, S.I. Branched fibres in old dystrophic mdx muscle are associated with mechanical weakening of the sarcolemma, abnormal Ca2+ transients and a breakdown of Ca2+ homeostasis during fatigue. Exp. Dermatol. 2010, 95, 641–656. [Google Scholar]
- de Lacy Costello, B.; Amann, A.; Al-Kateb, H.; Flynn, C.; Filipiak, W.; Khalid, T.; Osborne, D.; Ratcliffe, N.M. A review of the volatiles from the healthy human body. J. Breath Res. 2014, 8, 014001. [Google Scholar] [CrossRef] [PubMed]
- Opdyke, D.L.J. Fragrance raw materials monographs: α-cedrene. Food Cosmet. Toxicol. 1978, 16, 679–680. [Google Scholar] [CrossRef]
- Kang, W.; Zhang, K.; Tong, T.; Park, T. Improved glucose intolerance through a distinct mouse olfactory receptor 23-induced signaling pathway mediating glucose uptake In myotubes and adipocytes. Mol. Nutr. Food Res. 2020, 64, e1901329. [Google Scholar] [CrossRef]
- Vernochet, C.; Mourier, A.; Bezy, O.; Macotela, Y.; Boucher, J.; Rardin, M.J.; An, D.; Lee, K.Y.; Ilkayeva, O.R.; Zingaretti, C.M.; et al. Adipose-specific deletion of TFAM increases mitochondrial oxidation and protects mice against obesity and insulin resistance. Cell Metab. 2012, 16, 765–776. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pitman, J.L.; Wheeler, M.C.; Lloyd, D.J.; Walker, J.R.; Glynne, R.J.; Gekakis, N. A gain-of-function mutation in adenylate cyclase 3 protects mice from diet-induced obesity. PLoS ONE 2014, 9, e110226. [Google Scholar]
- Tong, T.; Shen, Y.; Lee, H.W.; Yu, R.; Park, T. Adenylyl cyclase 3 haploinsufficiency confers susceptibility to diet-induced obesity and insulin resistance in mice. Sci. Rep. 2016, 6, 34179. [Google Scholar] [CrossRef] [Green Version]
- Tong, T.; Yu, R.; Park, T. alpha-Cedrene protects rodents from high-fat diet-induced adiposity via adenylyl cyclase 3. Int. J. Obes. 2019, 43, 202–216. [Google Scholar] [CrossRef] [PubMed]
- Joo, J.I.; Kim, D.H.; Choi, J.W.; Yun, J.W. Proteomic analysis for antiobesity potential of capsaicin on white adipose tissue in rats fed with a high fat diet. J. Proteome Res. 2010, 9, 2977–2987. [Google Scholar] [CrossRef] [PubMed]
- Giusepponi, M.E.; Kern, M.; Chakaroun, R.; Wohland, T.; Kovacs, P.; Dietrich, A.; Schon, M.R.; Krohn, K.; Pucci, M.; Polidori, C.; et al. Gene expression profiling in adipose tissue of Sprague Dawley rats identifies olfactory receptor 984 as a potential obesity treatment target. Biochem. Biophys. Res. Commun. 2018, 505, 801–806. [Google Scholar] [CrossRef]
- Rasineni, K.; Casey, C.A. Molecular mechanism of alcoholic fatty liver. Indian J. Pharmacol. 2012, 44, 299–303. [Google Scholar] [CrossRef]
- Fichan, I.; Larroche, C.; Gros, J.B. Water solubility, vapor pressure, and activity coefficients of terpenes and terpenoids. J. Chem. Eng. Data 1999, 44, 56–62. [Google Scholar] [CrossRef]
- Furness, J.B.; Rivera, L.R.; Cho, H.J.; Bravo, D.M.; Callaghan, B. The gut as a sensory organ. Nat. Rev. Gastroenterol. Hepatol. 2013, 10, 729–740. [Google Scholar] [CrossRef]
- Gribble, F.M.; Reimann, F. Enteroendocrine cells: Chemosensors in the intestinal epithelium. Annu. Rev. Physiol. 2016, 78, 277–299. [Google Scholar] [CrossRef]
- Gershon, M.D. 5-Hydroxytryptamine (serotonin) in the gastrointestinal tract. Curr. Opin. Endocrinol. Diabetes Obes. 2013, 20, 14–21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Braun, T.; Voland, P.; Kunz, L.; Prinz, C.; Gratzl, M. Enterochromaffin cells of the human gut: Sensors for spices and odorants. Gastroenterology 2007, 132, 1890–1901. [Google Scholar] [CrossRef] [PubMed]
- Bellono, N.W.; Bayrer, J.R.; Leitch, D.B.; Castro, J.; Zhang, C.C.; O’Donnell, T.A.; Brierley, S.M.; Ingraham, H.A.; Julius, D. Enterochromaffin cells are gut chemosensors that couple to sensory neural pathways. Cell 2017, 170, 185–198.e116. [Google Scholar] [CrossRef] [Green Version]
- Han, Y.E.; Kang, C.W.; Oh, J.H.; Park, S.H.; Ku, C.R.; Cho, Y.H.; Lee, M.K.; Lee, E.J. Olfactory receptor OR51E1 mediates GLP-1 secretion in human and rodent enteroendocrine L cells. J. Endocr. Soc. 2018, 2, 1251–1258. [Google Scholar] [CrossRef] [PubMed]
- Rowland, I.; Gibson, G.; Heinken, A.; Scott, K.; Swann, J.; Thiele, I.; Tuohy, K. Gut microbiota functions: Metabolism of nutrients and other food components. Eur. J. Nutr. 2018, 57, 1–24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, K.S.; Lee, I.S.; Kim, K.H.; Park, J.; Kim, Y.; Choi, J.H.; Choi, J.S.; Jang, H.J. Activation of intestinal olfactory receptor stimulates glucagon-like peptide-1 secretion in enteroendocrine cells and attenuates hyperglycemia in type 2 diabetic mice. Sci. Rep. 2017, 7, 13978. [Google Scholar] [CrossRef] [Green Version]
- Kotlo, K.; Anbazhagan, A.N.; Priyamvada, S.; Jayawardena, D.; Kumar, A.; Chen, Y.; Xia, Y.; Finn, P.W.; Perkins, D.L.; Dudeja, P.K.; et al. The olfactory G protein-coupled receptor (Olfr-78/OR51E2) modulates the intestinal response to colitis. Am. J. Physiol. Cell Physiol. 2020, 318, C502–C513. [Google Scholar] [CrossRef]
- Shepard, B.D.; Cheval, L.; Peterlin, Z.; Firestein, S.; Koepsell, H.; Doucet, A.; Pluznick, J.L. A renal olfactory receptor aids in kidney glucose handling. Sci. Rep. 2016, 6, 35215. [Google Scholar] [CrossRef] [Green Version]
- Api, A.M.; Belmonte, F.; Belsito, D.; Botelho, D.; Bruze, M.; Burton, G.A., Jr.; Buschmann, J.; Dagli, M.L.; Date, M.; Dekant, W.; et al. RIFM fragrance ingredient safety assessment, isovaleric acid, CAS Registry Number 503-74-2. Food Chem. Toxicol. 2019, 130 (Suppl. S1), 110570. [Google Scholar] [CrossRef]
- Geng, R.; Kang, S.-G.; Huang, K.; Tong, T. Boosting the Photoaged Skin: The Potential Role of Dietary Components. Nutrients 2021, 13, 1691. [Google Scholar] [CrossRef]
- Denda, M.; Nakatani, M.; Ikeyama, K.; Tsutsumi, M.; Denda, S. Epidermal keratinocytes as the forefront of the sensory system. Exp. Dermatol. 2007, 16, 157–161. [Google Scholar] [CrossRef]
- Busse, D.; Kudella, P.; Gruning, N.M.; Gisselmann, G.; Stander, S.; Luger, T.; Jacobsen, F.; Steinstrasser, L.; Paus, R.; Gkogkolou, P.; et al. A synthetic sandalwood odorant induces wound-healing processes in human keratinocytes via the olfactory receptor OR2AT4. J. Investig. Dermatol. 2014, 134, 2823–2832. [Google Scholar] [CrossRef] [Green Version]
- Kim, B.E.; Goleva, E.; Hall, C.F.; Park, S.H.; Lee, U.H.; Brauweiler, A.M.; Streib, J.E.; Richers, B.N.; Kim, G.; Leung, D.Y.M. Skin wound healing is accelerated by a lipid mixture representing major lipid components of chamaecyparis obtusa plant extract. J. Investig. Dermatol. 2018, 138, 1176–1186. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, S.J.; Depoortere, I.; Hatt, H. Therapeutic potential of ectopic olfactory and taste receptors. Nat. Rev. Drug Discov. 2019, 18, 116–138. [Google Scholar] [CrossRef]
- Tsai, T.; Veitinger, S.; Peek, I.; Busse, D.; Eckardt, J.; Vladimirova, D.; Jovancevic, N.; Wojcik, S.; Gisselmann, G.; Altmuller, J.; et al. Two olfactory receptors-OR2A4/7 and OR51B5-differentially affect epidermal proliferation and differentiation. Exp. Dermatol. 2017, 26, 58–65. [Google Scholar] [CrossRef]
- Cheret, J.; Bertolini, M.; Ponce, L.; Lehmann, J.; Tsai, T.; Alam, M.; Hatt, H.; Paus, R. Olfactory receptor OR2AT4 regulates human hair growth. Nat. Commun. 2018, 9, 3624. [Google Scholar] [CrossRef]
- Gelis, L.; Jovancevic, N.; Veitinger, S.; Mandal, B.; Arndt, H.D.; Neuhaus, E.M.; Hatt, H. Functional characterization of the odorant receptor 51E2 in human melanocytes. J. Biol. Chem. 2016, 291, 17772–17786. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wojcik, S.; Weidinger, D.; Stander, S.; Luger, T.; Hatt, H.; Jovancevic, N. Functional characterization of the extranasal OR2A4/7 expressed in human melanocytes. Exp. Dermatol. 2018, 27, 1216–1223. [Google Scholar] [CrossRef] [PubMed]
- Tong, T.; Kim, N.; Park, T. Topical application of oleuropein induces anagen hair growth in telogen mouse skin. PLoS ONE 2015, 10, e0129578. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Esche, C.; de Benedetto, A.; Beck, L.A. Keratinocytes in atopic dermatitis: Inflammatory signals. Curr. Allergy Asthma Rep. 2004, 4, 276–284. [Google Scholar] [CrossRef] [PubMed]
- Duroux, R.; Mandeau, A.; Guiraudie-Capraz, G.; Quesnel, Y.; Loing, E. A Rose Extract Protects the Skin against Stress Mediators: A Potential Role of Olfactory Receptors. Molecules 2020, 25, 4743. [Google Scholar] [CrossRef]
- Duroux, R.; Mandeau, A.; Quesnel, Y.; Loing, E. Discovery of new olfactory receptors in human keratinocytes: A potential role on skin stress response. J. Investig. Dermatol. 2020, 140, S82. [Google Scholar] [CrossRef]
- Scognamiglio, J.; Jones, L.; Letizia, C.S.; Api, A.M. Fragrance material review on phenylethyl alcohol. Food Chem. Toxicol. 2012, 50 (Suppl. S2), S224–S239. [Google Scholar] [CrossRef]
- Jaeger, S.R.; McRae, J.F.; Bava, C.M.; Beresford, M.K.; Hunter, D.; Jia, Y.; Chheang, S.L.; Jin, D.; Peng, M.; Gamble, J.C.; et al. A Mendelian trait for olfactory sensitivity affects odor experience and food selection. Curr. Biol. 2013, 23, 1601–1605. [Google Scholar] [CrossRef] [Green Version]
- Langen, J.; Wegmann-Herr, P.; Schmarr, H.G. Quantitative determination of α-ionone, β-ionone, and β-damascenone and enantiodifferentiation of α-ionone in wine for authenticity control using multidimensional gas chromatography with tandem mass spectrometric detection. Anal. Bioanal. Chem. 2016, 408, 6483–6496. [Google Scholar] [CrossRef]
- Pan, S.; Conaway, S., Jr.; Deshpande, D.A. Mitochondrial regulation of airway smooth muscle functions in health and pulmonary diseases. Arch. Biochem. Biophys. 2019, 663, 109–119. [Google Scholar] [CrossRef] [PubMed]
- Pierre, S.; Eschenhagen, T.; Geisslinger, G.; Scholich, K. Capturing adenylyl cyclases as potential drug targets. Nat. Rev. Drug Discov. 2009, 8, 321–335. [Google Scholar] [CrossRef]
- Kalbe, B.; Knobloch, J.; Schulz, V.M.; Wecker, C.; Schlimm, M.; Scholz, P.; Jansen, F.; Stoelben, E.; Philippou, S.; Hecker, E.; et al. Olfactory receptors modulate physiological processes in human airway smooth muscle cells. Front. Physiol. 2016, 7, 339. [Google Scholar] [CrossRef] [Green Version]
- Aisenberg, W.H.; Huang, J.; Zhu, W.; Rajkumar, P.; Cruz, R.; Santhanam, L.; Natarajan, N.; Yong, H.M.; De Santiago, B.; Oh, J.J.; et al. Defining an olfactory receptor function in airway smooth muscle cells. Sci. Rep. 2016, 6, 38231. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gu, X.; Ben-Shahar, Y. Olfactory receptors in human airway epithelia. Methods Mol. Biol. 2013, 1003, 161–169. [Google Scholar]
- Api, A.M.; Belmonte, F.; Belsito, D.; Biserta, S.; Botelho, D.; Bruze, M.; Burton, G.A., Jr.; Buschmann, J.; Cancellieri, M.A.; Dagli, M.L.; et al. RIFM fragrance ingredient safety assessment, cinnamaldehyde, CAS Registry Number 104-55-2. Food Chem. Toxicol. 2019, 134 (Suppl. S1), 110710. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Lam, H.; Koziol-White, C.; Limjunyawong, N.; Kim, D.; Kim, N.; Karmacharya, N.; Rajkumar, P.; Firer, D.; Dalesio, N.M.; et al. The odorant receptor OR2W3 on airway smooth muscle evokes bronchodilation via a cooperative chemosensory tradeoff between TMEM16A and CFTR. Proc. Natl. Acad. Sci. USA 2020, 117, 28485–28495. [Google Scholar] [CrossRef] [PubMed]
- Sui, P.; Wiesner, D.L.; Xu, J.; Zhang, Y.; Lee, J.; Van Dyken, S.; Lashua, A.; Yu, C.; Klein, B.S.; Locksley, R.M.; et al. Pulmonary neuroendocrine cells amplify allergic asthma responses. Science 2018, 360, eaan8546. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, J.J.; Tay, H.L.; Plank, M.; Essilfie, A.T.; Hansbro, P.M.; Foster, P.S.; Yang, M. Activation of olfactory receptors on mouse pulmonary macrophages promotes monocyte chemotactic protein-1 production. PLoS ONE 2013, 8, e80148. [Google Scholar] [CrossRef]
- Chakraborty, S.; Dakle, P.; Sinha, A.; Vishweswaraiah, S.; Nagori, A.; Salimath, S.; Prakash, Y.S.; Lodha, R.; Kabra, S.K.; Ghosh, B.; et al. Genetic variations in olfactory receptor gene OR2AG2 in a large multigenerational family with asthma. Sci. Rep. 2019, 9, 19029. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, S.H.; Yoon, Y.C.; Lee, A.S.; Kang, N.; Koo, J.; Rhyu, M.R.; Park, J.H. Expression of human olfactory receptor 10J5 in heart aorta, coronary artery, and endothelial cells and its functional role in angiogenesis. Biochem. Biophys. Res. Commun. 2015, 460, 404–408. [Google Scholar] [CrossRef]
- Li, W.H. Functional analysis of islet cells in vitro, in situ, and in vivo. Semin. Cell Dev. Biol. 2020, 103, 14–19. [Google Scholar] [CrossRef]
- Wang, X.; Kang, J.; Liu, Q.; Tong, T.; Quan, H. Fighting diabetes mellitus: Pharmacological and non-pharmacological approaches. Curr. Pharm. Des. 2020, 26, 4992–5001. [Google Scholar] [CrossRef]
- Api, A.M.; Belsito, D.; Biserta, S.; Botelho, D.; Bruze, M.; Burton, G.A., Jr.; Buschmann, J.; Cancellieri, M.A.; Dagli, M.L.; Date, M.; et al. RIFM fragrance ingredient safety assessment, octanoic acid, CAS Registry Number 124-07-2. Food Chem. Toxicol. 2020, 138 (Suppl. S1), 111271. [Google Scholar] [CrossRef]
- Lee, N.; Jae, Y.; Kim, M.; Cho, T.; Lee, C.; Hong, Y.R.; Hyeon, D.Y.; Ahn, S.; Kwon, H.; Kim, K.; et al. A pathogen-derived metabolite induces microglial activation via odorant receptors. FEBS J. 2020, 287, 3841–3870. [Google Scholar] [CrossRef] [PubMed]
- Ahrén, B. Islet G protein-coupled receptors as potential targets for treatment of type 2 diabetes. Nat. Rev. Drug Discov. 2009, 8, 369–385. [Google Scholar] [CrossRef]
- Munakata, Y.; Yamada, T.; Imai, J.; Takahashi, K.; Tsukita, S.; Shirai, Y.; Kodama, S.; Asai, Y.; Sugisawa, T.; Chiba, Y.; et al. Olfactory receptors are expressed in pancreatic beta-cells and promote glucose-stimulated insulin secretion. Sci. Rep. 2018, 8, 1499. [Google Scholar] [CrossRef] [PubMed]
- Leem, J.; Shim, H.M.; Cho, H.; Park, J.H. Octanoic acid potentiates glucose-stimulated insulin secretion and expression of glucokinase through the olfactory receptor in pancreatic beta-cells. Biochem. Biophys. Res. Commun. 2018, 503, 278–284. [Google Scholar] [CrossRef] [PubMed]
- Kalbe, B.; Schlimm, M.; Mohrhardt, J.; Scholz, P.; Jansen, F.; Hatt, H.; Osterloh, S. Helional induces Ca2+ decrease and serotonin secretion of QGP-1 cells via a PKG-mediated pathway. J. Mol. Endocrinol. 2016, 57, 201–210. [Google Scholar] [CrossRef] [Green Version]
- Paulmann, N.; Grohmann, M.; Voigt, J.P.; Bert, B.; Vowinckel, J.; Bader, M.; Skelin, M.; Jevsek, M.; Fink, H.; Rupnik, M.; et al. Intracellular serotonin modulates insulin secretion from pancreatic beta-cells by protein serotonylation. PLoS Biol. 2009, 7, e1000229. [Google Scholar] [CrossRef] [Green Version]
- Gong, L.; Chen, Q.; Gu, X.; Li, S. Expression and identification of olfactory receptors in sciatic nerve and dorsal root ganglia of rats. Neurosci. Lett. 2015, 600, 171–175. [Google Scholar] [CrossRef]
- Garcia-Esparcia, P.; Schluter, A.; Carmona, M.; Moreno, J.; Ansoleaga, B.; Torrejon-Escribano, B.; Gustincich, S.; Pujol, A.; Ferrer, I. Functional genomics reveals dysregulation of cortical olfactory receptors in Parkinson disease: Novel putative chemoreceptors in the human brain. J. Neuropathol. Exp. Neurol. 2013, 72, 524–539. [Google Scholar] [CrossRef] [Green Version]
- Ansoleaga, B.; Garcia-Esparcia, P.; Llorens, F.; Moreno, J.; Aso, E.; Ferrer, I. Dysregulation of brain olfactory and taste receptors in AD, PSP and CJD, and AD-related model. Neuroscience 2013, 248, 369–382. [Google Scholar] [CrossRef]
- Cho, T.; Lee, C.; Lee, N.; Hong, Y.R.; Koo, J. Small-chain fatty acid activates astrocytic odorant receptor Olfr920. Biochem. Biophys. Res. Commun. 2019, 510, 383–387. [Google Scholar] [CrossRef]
- Rodriguez, M.; Siwko, S.; Zeng, L.; Li, J.; Yi, Z.; Liu, M. Prostate-specific G-protein-coupled receptor collaborates with loss of PTEN to promote prostate cancer progression. Oncogene 2016, 35, 1153–1162. [Google Scholar] [CrossRef]
- Sanz, G.; Leray, I.; Dewaele, A.; Sobilo, J.; Lerondel, S.; Bouet, S.; Grebert, D.; Monnerie, R.; Pajot-Augy, E.; Mir, L.M. Promotion of cancer cell invasiveness and metastasis emergence caused by olfactory receptor stimulation. PLoS ONE 2014, 9, e85110. [Google Scholar] [CrossRef]
- Abaffy, T.; Bain, J.R.; Muehlbauer, M.J.; Spasojevic, I.; Lodha, S.; Bruguera, E.; O’Neal, S.K.; Kim, S.Y.; Matsunami, H. A Testosterone Metabolite 19-Hydroxyandrostenedione Induces Neuroendocrine Trans-Differentiation of Prostate Cancer Cells via an Ectopic Olfactory Receptor. Front. Oncol. 2018, 8, 162. [Google Scholar] [CrossRef] [Green Version]
- Spehr, J.; Gelis, L.; Osterloh, M.; Oberland, S.; Hatt, H.; Spehr, M.; Neuhaus, E.M. G protein-coupled receptor signaling via Src kinase induces endogenous human transient receptor potential vanilloid type 6 (TRPV6) channel activation. J. Biol. Chem. 2011, 286, 13184–13192. [Google Scholar] [CrossRef] [Green Version]
- Gelis, L.; Jovancevic, N.; Bechara, F.G.; Neuhaus, E.M.; Hatt, H. Functional expression of olfactory receptors in human primary melanoma and melanoma metastasis. Exp. Dermatol. 2017, 26, 569–576. [Google Scholar] [CrossRef]
- Manteniotis, S.; Wojcik, S.; Brauhoff, P.; Mollmann, M.; Petersen, L.; Gothert, J.R.; Schmiegel, W.; Duhrsen, U.; Gisselmann, G.; Hatt, H. Functional characterization of the ectopically expressed olfactory receptor 2AT4 in human myelogenous leukemia. Cell Death Discov. 2016, 2, 15070. [Google Scholar] [CrossRef]
- Weber, L.; Al-Refae, K.; Ebbert, J.; Jagers, P.; Altmuller, J.; Becker, C.; Hahn, S.; Gisselmann, G.; Hatt, H. Activation of odorant receptor in colorectal cancer cells leads to inhibition of cell proliferation and apoptosis. PLoS ONE 2017, 12, e0172491. [Google Scholar] [CrossRef] [Green Version]
- Massberg, D.; Simon, A.; Haussinger, D.; Keitel, V.; Gisselmann, G.; Conrad, H.; Hatt, H. Monoterpene (-)-citronellal affects hepatocarcinoma cell signaling via an olfactory receptor. Arch. Biochem. Biophys. 2015, 566, 100–109. [Google Scholar] [CrossRef] [PubMed]
- Weber, L.; Schulz, W.A.; Philippou, S.; Eckardt, J.; Ubrig, B.; Hoffmann, M.J.; Tannapfel, A.; Kalbe, B.; Gisselmann, G.; Hatt, H. Characterization of the olfactory receptor OR10H1 in human urinary bladder cancer. Front. Physiol. 2018, 9, 456. [Google Scholar] [CrossRef]
- Kalbe, B.; Schulz, V.M.; Schlimm, M.; Philippou, S.; Jovancevic, N.; Jansen, F.; Scholz, P.; Lubbert, H.; Jarocki, M.; Faissner, A.; et al. Helional-induced activation of human olfactory receptor 2J3 promotes apoptosis and inhibits proliferation in a non-small-cell lung cancer cell line. Eur. J. Cell Biol. 2017, 96, 34–46. [Google Scholar] [CrossRef]
- Massberg, D.; Jovancevic, N.; Offermann, A.; Simon, A.; Baniahmad, A.; Perner, S.; Pungsrinont, T.; Luko, K.; Philippou, S.; Ubrig, B.; et al. The activation of OR51E1 causes growth suppression of human prostate cancer cells. Oncotarget 2016, 7, 48231–48249. [Google Scholar] [CrossRef] [Green Version]
- Giandomenico, V.; Cui, T.; Grimelius, L.; Oberg, K.; Pelosi, G.; Tsolakis, A.V. Olfactory receptor 51E1 as a novel target for diagnosis in somatostatin receptor-negative lung carcinoids. J. Mol. Endocrinol. 2013, 51, 277–286. [Google Scholar] [CrossRef] [Green Version]
- Manteniotis, S.; Wojcik, S.; Gothert, J.R.; Durig, J.; Duhrsen, U.; Gisselmann, G.; Hatt, H. Deorphanization and characterization of the ectopically expressed olfactory receptor OR51B5 in myelogenous leukemia cells. Cell Death Discov. 2016, 2, 16010. [Google Scholar] [CrossRef] [Green Version]
- Li, M.; Wang, X.; Ma, R.R.; Shi, D.B.; Wang, Y.W.; Li, X.M.; He, J.Y.; Wang, J.; Gao, P. The olfactory receptor family 2, subfamily T, member 6 (OR2T6) is involved in breast cancer progression via initiating epithelial-mesenchymal transition and MAPK/ERK pathway. Front. Oncol. 2019, 9, 1210. [Google Scholar] [CrossRef]
- Morita, R.; Hirohashi, Y.; Torigoe, T.; Ito-Inoda, S.; Takahashi, A.; Mariya, T.; Asanuma, H.; Tamura, Y.; Tsukahara, T.; Kanaseki, T.; et al. Olfactory receptor family 7 subfamily c member 1 is a novel marker of colon cancer-initiating cells and is a potent target of immunotherapy. Clin. Cancer Res. 2016, 22, 3298–3309. [Google Scholar] [CrossRef] [Green Version]
- Weber, L.; Massberg, D.; Becker, C.; Altmuller, J.; Ubrig, B.; Bonatz, G.; Wolk, G.; Philippou, S.; Tannapfel, A.; Hatt, H.; et al. Olfactory receptors as biomarkers in human breast carcinoma tissues. Front. Oncol. 2018, 8, 33. [Google Scholar] [CrossRef] [Green Version]
- Moein, S.T.; Hashemian, S.M.; Mansourafshar, B.; Khorram-Tousi, A.; Tabarsi, P.; Doty, R.L. Smell dysfunction: A biomarker for COVID-19. Int. Forum. Allergy Rhinol. 2020, 10, 944–950. [Google Scholar] [CrossRef]
- Abaffy, T.; Matsunami, H.; Luetje, C.W. Functional analysis of a mammalian odorant receptor subfamily. J. Neurochem. 2006, 97, 1506–1518. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matarazzo, V.; Clot-Faybesse, O.; Marcet, B.; Guiraudie-Capraz, G.; Atanasova, B.; Devauchelle, G.; Cerutti, M.; Etievant, P.; Ronin, C. Functional characterization of two human olfactory receptors expressed in the baculovirus Sf9 insect cell system. Chem. Senses 2005, 30, 195–207. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kajiya, K.; Inaki, K.; Tanaka, M.; Haga, T.; Kataoka, H.; Touhara, K. Molecular bases of odor discrimination: Reconstitution of olfactory receptors that recognize overlapping sets of odorants. J. Neurosci. 2001, 21, 6018–6025. [Google Scholar] [CrossRef]
- Saito, H.; Kubota, M.; Roberts, R.W.; Chi, Q.; Matsunami, H. RTP family members induce functional expression of mammalian odorant receptors. Cell 2004, 119, 679–691. [Google Scholar] [CrossRef] [Green Version]
- Peterlin, Z.; Firestein, S.; Rogers, M.E. The state of the art of odorant receptor deorphanization: A report from the orphanage. J. Gen. Physiol. 2014, 143, 527–542. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhuang, H.; Matsunami, H. Evaluating cell-surface expression and measuring activation of mammalian odorant receptors in heterologous cells. Nat. Protoc. 2008, 3, 1402–1413. [Google Scholar] [CrossRef] [Green Version]
- Dunkel, A.; Steinhaus, M.; Kotthoff, M.; Nowak, B.; Krautwurst, D.; Schieberle, P.; Hofmann, T. Nature’s chemical signatures in human olfaction: A foodborne perspective for future biotechnology. Angew. Chem. Int. Ed. Engl. 2014, 53, 7124–7143. [Google Scholar] [CrossRef]
- Saito, H.; Chi, Q.; Zhuang, H.; Matsunami, H.; Mainland, J.D. Odor coding by a Mammalian receptor repertoire. Sci. Signal. 2009, 2, ra9. [Google Scholar] [CrossRef] [Green Version]
- Mainland, J.D.; Keller, A.; Li, Y.R.; Zhou, T.; Trimmer, C.; Snyder, L.L.; Moberly, A.H.; Adipietro, K.A.; Liu, W.L.; Zhuang, H.; et al. The missense of smell: Functional variability in the human odorant receptor repertoire. Nat. Neurosci. 2014, 17, 114–120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adipietro, K.A.; Mainland, J.D.; Matsunami, H. Functional evolution of mammalian odorant receptors. PLoS Genet. 2012, 8, e1002821. [Google Scholar] [CrossRef] [Green Version]
- Geithe, C.; Noe, F.; Kreissl, J.; Krautwurst, D. The broadly tuned odorant receptor OR1A1 is highly selective for 3-methyl-2,4-nonanedione, a key food odorant in aged wines, tea, and other foods. Chem. Senses 2017, 42, 181–193. [Google Scholar] [CrossRef]
- Sato-Akuhara, N.; Horio, N.; Kato-Namba, A.; Yoshikawa, K.; Niimura, Y.; Ihara, S.; Shirasu, M.; Touhara, K. Ligand specificity and evolution of mammalian musk odor receptors: Effect of single receptor deletion on odor detection. J. Neurosci. 2016, 36, 4482–4491. [Google Scholar] [CrossRef] [Green Version]
- Schmiedeberg, K.; Shirokova, E.; Weber, H.P.; Schilling, B.; Meyerhof, W.; Krautwurst, D. Structural determinants of odorant recognition by the human olfactory receptors OR1A1 and OR1A2. J. Struct. Biol. 2007, 159, 400–412. [Google Scholar] [CrossRef]
- Ahmed, L.; Zhang, Y.; Block, E.; Buehl, M.; Corr, M.J.; Cormanich, R.A.; Gundala, S.; Matsunami, H.; O’Hagan, D.; Ozbil, M.; et al. Molecular mechanism of activation of human musk receptors OR5AN1 and OR1A1 by (R)-muscone and diverse other musk-smelling compounds. Proc. Natl. Acad. Sci. USA 2018, 115, E3950–E3958. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Veithen, A.; Wilin, F.; Philippeau, M.; Chatelain, P. OR1D2 is a broadly tuned human olfactory receptor. Chem. Senses 2015, 40, 262–263. [Google Scholar]
- Sanz, G.; Schlegel, C.; Pernollet, J.C.; Briand, L. Comparison of odorant specificity of two human olfactory receptors from different phylogenetic classes and evidence for antagonism. Chem. Senses 2005, 30, 69–80. [Google Scholar] [CrossRef]
- Charlier, L.; Topin, J.; Ronin, C.; Kim, S.K.; Goddard, W.A., 3rd; Efremov, R.; Golebiowski, J. How broadly tuned olfactory receptors equally recognize their agonists. Human OR1G1 as a test case. Cell Mol. Life Sci. 2012, 69, 4205–4213. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Kristeller, D.C.; do Nascimento, J.B.; Galante, P.A.; Malnic, B. Identification of agonists for a group of human odorant receptors. Front. Pharmacol. 2015, 6, 35. [Google Scholar] [CrossRef] [Green Version]
- Topin, J.; de March, C.A.; Charlier, L.; Ronin, C.; Antonczak, S.; Golebiowski, J. Discrimination between olfactory receptor agonists and non-agonists. Chemistry 2014, 20, 10227–10230. [Google Scholar] [CrossRef] [PubMed]
- Neuhaus, E.M.; Mashukova, A.; Zhang, W.; Barbour, J.; Hatt, H. A specific heat shock protein enhances the expression of mammalian olfactory receptor proteins. Chem. Senses 2006, 31, 445–452. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mcrae, J.F.; Mainland, J.D.; Jaeger, S.R.; Adipietro, K.A.; Matsunami, H.; Newcomb, R.D. Genetic variation in the odorant receptor OR2J3 is associated with the ability to detect the “grassy” smelling odor, cis-3-hexen-1-ol. Chem. Senses 2012, 37, 585–593. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Noe, F.; Polster, J.; Geithe, C.; Kotthoff, M.; Schieberle, P.; Krautwurst, D. OR2M3: A highly specific and narrowly tuned human odorant receptor for the sensitive detection of onion key food odorant 3-mercapto-2-methylpentan-1-ol. Chem. Senses 2017, 42, 195–210. [Google Scholar] [CrossRef] [PubMed]
- Kotthoff, M.; Schieberle, P.; Krautwurst, D. Human olfactory receptors for thiol key food odorants. In Wartburg Symposium on Flavor Chemistry & Biology, 9th ed.; Technische Universitaet Muenchen: Eisenach, Germany, 2010; pp. 144–147. [Google Scholar]
- Yasi, E.A.; Eisen, S.L.; Wang, H.; Sugianto, W.; Minniefield, A.R.; Hoover, K.A.; Branham, P.J.; Peralta-Yahya, P. Rapid deorphanization of human olfactory receptors in yeast. Biochemistry 2019, 58, 2160–2166. [Google Scholar] [CrossRef]
- Li, S.; Ahmed, L.; Zhang, R.; Pan, Y.; Matsunami, H.; Burger, J.L.; Block, E.; Batista, V.S.; Zhuang, H. Smelling sulfur: Copper and silver regulate the response of human odorant receptor OR2T11 to low-molecular-weight thiols. J. Am. Chem. Soc. 2016, 138, 13281–13288. [Google Scholar] [CrossRef]
- Jacquier, V.; Pick, H.; Vogel, H. Characterization of an extended receptive ligand repertoire of the human olfactory receptor OR17-40 comprising structurally related compounds. J. Neurochem. 2006, 97, 537–544. [Google Scholar] [CrossRef] [Green Version]
- Wetzel, C.H.; Oles, M.; Wellerdieck, C.; Kuczkowiak, M.; Gisselmann, G.; Hatt, H. Specificity and sensitivity of a human olfactory receptor functionally expressed in human embryonic kidney 293 cells and Xenopus Laevis oocytes. J. Neurosci. 1999, 19, 7426–7433. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, W.; Ho, L.; Varghese, M.; Yemul, S.; Dams-O’Connor, K.; Gordon, W.; Knable, L.; Freire, D.; Haroutunian, V.; Pasinetti, G.M. Decreased level of olfactory receptors in blood cells following traumatic brain injury and potential association with tauopathy. J. Alzheimers Dis. 2013, 34, 417–429. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shirasu, M.; Yoshikawa, K.; Takai, Y.; Nakashima, A.; Takeuchi, H.; Sakano, H.; Touhara, K. Olfactory receptor and neural pathway responsible for highly selective sensing of musk odors. Neuron 2014, 81, 165–178. [Google Scholar] [CrossRef] [Green Version]
- Veithen, A.; Philippeau, M.; Wilkin, F.; Chatelain, P. OR5D3P, a pseudogene with a functional activity potential. Chem. Senses 2009, 34, A58. [Google Scholar]
- Marcinek, P.; Haag, F.; Geithe, C.; Krautwurst, D. An evolutionary conserved olfactory receptor for foodborne and semiochemical alkylpyrazines. FASEB J. 2021, 35, e21638. [Google Scholar] [CrossRef]
- Keller, A.; Zhuang, H.; Chi, Q.; Vosshall, L.B.; Matsunami, H. Genetic variation in a human odorant receptor alters odour perception. Nature 2007, 449, 468–472. [Google Scholar] [CrossRef]
- Keto, A.; Saito, N. Method for Identifying a Malodor Inhibitor. WO Patent 2012,169,644, 6 May 2012. [Google Scholar]
- Mainland, J.D.; Li, Y.R.; Zhou, T.; Liu, W.L.; Matsunami, H. Human olfactory receptor responses to odorants. Sci. Data 2015, 2, 150002. [Google Scholar] [CrossRef] [Green Version]
- Menashe, I.; Abaffy, T.; Hasin, Y.; Goshen, S.; Yahalom, V.; Luetje, C.W.; Lancet, D. Genetic elucidation of human hyperosmia to isovaleric acid. PLoS Biol. 2007, 5, e284. [Google Scholar] [CrossRef]
- Pfister, P.; Rogers, M.E.; Parris, K.J. Methods of Identifying, Isolating and Using Odorant and Aroma Receptors. U.S. Patent 20180208637A1, 1 January 2015. [Google Scholar]
- Jovancevic, N.; Dendorfer, A.; Matzkies, M.; Kovarova, M.; Heckmann, J.C.; Osterloh, M.; Boehm, M.; Weber, L.; Nguemo, F.; Semmler, J.; et al. Medium-chain fatty acids modulate myocardial function via a cardiac odorant receptor. Basic Res. Cardiol. 2017, 112, 13. [Google Scholar] [CrossRef] [Green Version]
- Fujita, Y.; Takahashi, T.; Suzuki, A.; Kawashima, K.; Nara, F.; Koishi, R. Deorphanization of Dresden G protein-coupled receptor for an odorant receptor. J. Recept. Signal Transduct. Res. 2007, 27, 323–334. [Google Scholar] [CrossRef]
- Sanz, G.; Leray, I.; Grebert, D.; Antoine, S.; Acquistapace, A.; Muscat, A.; Boukadiri, A.; Mir, L.M. Structurally related odorant ligands of the olfactory receptor OR51E2 differentially promote metastasis emergence and tumor growth. Oncotarget 2017, 8, 4330–4341. [Google Scholar] [CrossRef] [Green Version]
- Kato, A.; Saito, N.; Wakisaka, E. Method for Searching for Malodor Control Agent, Malodor Control Agent, and Malodor Control Metho. U.S. Patent 20130,216,492, 3 August 2013. [Google Scholar]
- Chatelain, P.; Veithen, A. Olfactory Receptors Involved in the Perception of Sweat Carboxylic Acids and the Use Thereof. U.S. Patent 20130,336,910 A1, 19 December 2013. [Google Scholar]
- Pichavant, C.; Burkholder, T.J.; Pavlath, G.K. Decrease of myofiber branching via muscle-specific expression of the olfactory receptor mOR23 in dystrophic muscle leads to protection against mechanical stress. Skelet Muscle 2016, 6, 2. [Google Scholar] [CrossRef] [Green Version]
- Kalra, S.; Mittal, A.; Bajoria, M.; Mishra, T.; Maryam, S.; Sengupta, D.; Ahuja, G. Challenges and possible solutions for decoding extranasal olfactory receptors. FEBS J. 2021, 288, 4230–4241. [Google Scholar] [CrossRef] [PubMed]
- Di Pizio, A.; Behrens, M.; Krautwurst, D. Beyond the flavour: The potential druggability of chemosensory G protein-coupled receptors. Int. J. Mol. Sci. 2019, 20, 1402. [Google Scholar] [CrossRef] [Green Version]






| OR | Ligands | Refs |
|---|---|---|
| OR1A1 | Allyl heptanoate | [135] |
| Allyl phenyl acetate | [135,136,137] | |
| trans-Anethole | [138] | |
| Benzophenone | [135] | |
| Benzyl acetate | [135] | |
| Bourgeonal | [63] | |
| Cosmone | [139] | |
| Celestolide | [139] | |
| Citral | [137,140] | |
| (+)-Carvone | [135,136,137,138] | |
| (-)-Carvone | [135,138] | |
| 4-Chromanone | [135] | |
| (S)-(-)-citronellal | [140] | |
| (S)-(-)-Citronellol | [140] | |
| (R)-(+)-Citronellol | [140] | |
| (-)-β-Citronellol | [135] | |
| (-)-Carveol | [140] | |
| Dihydrojasmone | [135] | |
| (+)-Dihydrocarvone | [135] | |
| 4-Decenal | [140] | |
| Estragole | [138] | |
| Ethyl cyclohexanecarboxylate | [138] | |
| Ethyl hexanoate | [138] | |
| Ethylphenyl acetate | [138] | |
| Geraniol | [63,135,140] | |
| Helional | [63,137,140] | |
| Heptanal | [140] | |
| Hydroxy-citronellal | [140] | |
| 3-Heptanone | [135] | |
| (S)-(-)-Limonene | [138] | |
| (R)-(+)-Limonene | [137,138] | |
| Musk ambrette | [141] | |
| Musk xylene | [141] | |
| Musk tibetene | [141] | |
| Muscone | [139] | |
| Musk xylol | [139] | |
| Muscenone | [139] | |
| (-)-Menthone | [138] | |
| (+)-Menthone | [138] | |
| 3-Mercaptohexyl acetate | [138] | |
| 3-Methyl-2,4-nonanedione | [138] | |
| 2-Nonanone | [138] | |
| Nonanal | [140] | |
| Nonanethiol | [135] | |
| (R/S)-γ-Nonalactone | [138] | |
| Octanal | [140] | |
| Octanethiol | [135] | |
| Octanol | [140] | |
| Quinoline | [137] | |
| (R/S)-Octen-3-ol | [138] | |
| 2-Octanone | [135] | |
| 3-Octanone | [135] | |
| 2-Pentylpyridine | [138] | |
| 2-Phenylethyl acetate | [138] | |
| 2-Phenylethanethiol | [138] | |
| OR1A2 | Citral | [140] |
| (-)-Carveol | [140] | |
| (R)-(+)-Citronellol | [140] | |
| (S)-(-)-Citronellal | [140] | |
| 4-Decenal | [140] | |
| Geraniol | [140] | |
| Helional | [140] | |
| Heptanal | [140] | |
| Hydroxy-citronellal | [140] | |
| Nonanal | [140] | |
| Octanal | [140] | |
| Octanol | [140] | |
| OR1C1 | Androstenone | [137] |
| Coumarin | [30,137] | |
| Linalool | [136] | |
| Nonanoic acid | [137] | |
| OR1D2 | Anisyl acetate | [142] |
| Allyl cyclohexylpropionate | [142] | |
| Benzyl isobutyrate | [142] | |
| Benzyl acetone | [142] | |
| Bourgeonal | [5,30,63] | |
| Benzyl propionate | [142] | |
| Benzyl acetate | [142] | |
| Benzyl butyrate | [142] | |
| Canthoxal | [5] | |
| Cyclamal | [5] | |
| Citral dimethyl acetal | [142] | |
| Coranol | [142] | |
| Citronellol | [142] | |
| Cassione | [142] | |
| Citral | [142] | |
| Citronellyl nitrile | [142] | |
| Cinnamic alcohol | [142] | |
| Citronellyl oxyacetaldehyde | [142] | |
| (S)-(-)-Citronellal | [142] | |
| Cinnamyl nitrile | [142] | |
| Clonal | [142] | |
| Dihydromyrcenol | [142] | |
| Dimethyl ethyl phenyl carbinol | [142] | |
| Dihydroisojasmonate | [142] | |
| 2,6-dimethyl-7-octen-2-ol | [142] | |
| (E),(E)-2,4-Decadienal | [142] | |
| δ-Dodecalactone | [142] | |
| γ-Dodecalactone | [142] | |
| 9-Decen-1-ol | [142] | |
| Diethyleneglycol hexyl ether | [142] | |
| 5-Decanol | [142] | |
| γ-Decalactone | [142] | |
| δ-2-Decenolactone | [142] | |
| δ-Decalactone | [142] | |
| Ethyl heptanone | [142] | |
| Ethyl cinnamate | [142] | |
| Ethyl p-anisate | [142] | |
| Ethyl linalool | [142] | |
| Ethylene Glycol Monophenoxyacetate | [142] | |
| Empetal | [142] | |
| Ethyl phenyl glycidate | [142] | |
| Florymoss | [142] | |
| Frutonile | [142] | |
| Floralozone | [5] | |
| Geraniol | [142] | |
| Geranyl propionate | [142] | |
| Geranyl butyrate | [142] | |
| Geranyl acetate | [142] | |
| Hypo-lem | [142] | |
| Heptanal | [142] | |
| Hepto | [142] | |
| Iso jasmone | [142] | |
| Jasmatone | [142] | |
| Linalool | [142] | |
| Lilial | [5] | |
| Methyl naphthyl ketone | [142] | |
| Mefrosol | [142] | |
| Methyl-trans-cinnamate | [142] | |
| Myrcenol | [142] | |
| Milk lactone 2067 | [142] | |
| Methyl tuberate | [142] | |
| 7-Methylindole | [142] | |
| β-Methylphenylethylamine | [142] | |
| 5-Methylindole | [142] | |
| para-Methoxyacetophenone | [142] | |
| 1-Methylindole | [142] | |
| Methyl nicotinate | [142] | |
| 3-Methyl-3-nonanol | [142] | |
| 1-Nonanol | [142] | |
| Nerolidyl acetate | [142] | |
| Nonalactone | [142] | |
| cis-6-Nonen-1-ol | [142] | |
| trans-2-Nonenal | [142] | |
| Neryl acetate | [142] | |
| δ-Nonalactone | [142] | |
| Nonanal | [142] | |
| Octanal | [142] | |
| 3-Octyl acetate | [142] | |
| Quintone | [142] | |
| cis-5-Octen-1-ol | [142] | |
| 7-Octen1-ol | [142] | |
| 1-Octen-3-ol | [142] | |
| 1-Octyl-2-pyrrolidone | [142] | |
| Petiole | [142] | |
| Pivarose | [142] | |
| Phenyl acetaldehyde | [5] | |
| 3-Phenylbutyraldehyde | [5] | |
| 3-Phenylpropionic aldehyde | [5] | |
| 4-Phenylbutyraldehyde | [5] | |
| Phenyl ethyl isovalerate | [142] | |
| Phenyl ethyl acetate | [142] | |
| β-Phenoxy-ethyl-isobutyrate | [142] | |
| Rosaphen | [142] | |
| Tetrahydromyrcenol | [142] | |
| Tetrahydrolinalool | [142] | |
| (4-Tert-butylphenoxy) acetaldehyde | [5] | |
| Tetrahydrogeraniol | [142] | |
| δ-Tridecalactone | [142] | |
| Tetrahydro citral | [142] | |
| δ-Tetradecalactone | [142] | |
| δ-Undecalactone | [142] | |
| Undecanal (antagonist) | [5] | |
| γ-Undecalactone | [142] | |
| Undecene-2-nitrile | [142] | |
| Violet nitrile | [142] | |
| Zinarine | [142] | |
| OR1E3 | Acetophenone | [63,129] |
| OR1G1 | Acetophenone | [63,143] |
| Benzaldehyde | [143] | |
| Benzothiazol | [143] | |
| β-Ionone | [63] | |
| Camphor | [143,144] | |
| Capric acid | [143] | |
| Cinnamaldehyde | [145] | |
| Citral | [143] | |
| Coumarin | [143] | |
| (+/-)-Citronellal | [143] | |
| Decanal | [143] | |
| 9-Decen-1-ol | [144] | |
| 1-Decanol | [143] | |
| γ-Decalactone | [143] | |
| 1-Dodecanol | [143] | |
| Ethyl butyrate | [143] | |
| Ethyl decanoate | [143] | |
| Ethyl isobutyrate | [143] | |
| Ethy-2-methyl propanoate | [134,143] | |
| Ethyl nonanoate | [143] | |
| Ethyl octanoate | [143] | |
| Ethyl vanillin | [143] | |
| Eugenyl acetate | [145] | |
| 2-Ethyl-1-hexanol | [143] | |
| Floralozone | [145] | |
| Geraniol | [63,143] | |
| Guaiacol | [143] | |
| Hedione | [143] | |
| Heptanal | [143] | |
| Hexanal | [143] | |
| 1-Heptanol | [143] | |
| 1-Hexanol | [143] | |
| 3-Hydroxybutan-2-one | [143] | |
| Isoamyl acetate | [63,129,143] | |
| 2-Isobutyl-3-methoxypyrazine | [143] | |
| Jasmonyl | [145] | |
| Lauric aldehyde | [143] | |
| Limonene | [143] | |
| Lyral | [143] | |
| Maltol | [143] | |
| Maltyl isobutyrate | [145] | |
| Manzanate | [145] | |
| Menthol | [143] | |
| Methyl decanoate | [143] | |
| Methyl nonanoate | [143] | |
| Methyl octanoate | [143] | |
| 2-Methyl pyrazine | [143] | |
| Nonanal | [143] | |
| Nonanoic acid | [143] | |
| 2-Nonanol | [143] | |
| 2-Nonanone | [143] | |
| 3-Nonanone | [143] | |
| 1-Nonanol | [143,144] | |
| Octanal | [143] | |
| Octanol | [143] | |
| 3-Octanol | [143] | |
| 4-Octanol | [143] | |
| 2-Octanol | [143] | |
| Phenylmethanol | [143] | |
| Piperonyl acetone | [143] | |
| Pyrazine | [143] | |
| Pyridin | [143] | |
| Quinoline | [143] | |
| Safrole | [143] | |
| S-methylthio butanoate | [143] | |
| Thiazol | [143] | |
| Thymol | [63,143] | |
| Trans-anethol | [143] | |
| Tridecanal | [143] | |
| 2-Undecanone | [146] | |
| Vanillin | [143] | |
| OR1L3 | α-Damascone | [145] |
| Vanilin | [145] | |
| OR2A25 | Geranyl acetate | [136,137] |
| Quinoline | [137] | |
| OR2A4/7 | Cyclohexyl salicylate | [76] |
| OR2AG1 | Amylbutyrate | [147] |
| OR2AG2 | Benzyl acetone | [83] |
| Citronellol | [83] | |
| Cis-3-Hexenol | [83] | |
| α-Cinnamyl alcohol | [83] | |
| Geraniol | [83] | |
| Linalool | [83] | |
| Nerol | [83] | |
| Phenyl ethyl alcohol | [83] | |
| Phenyl propyl alcohol | [83] | |
| OR2AT4 | Brahmanol | [73] |
| Oxyphenylon(Antagonist) | [73] | |
| Phenirat (Antagonist) | [73] | |
| Sandalore | [73] | |
| OR2B11 | Cinnamaldehyde | [136] |
| Coumarin | [136,137] | |
| (R)-(+)-Limonene | [137] | |
| Quinoline | [137] | |
| OR2B3 | Eugenyl acetate | [145] |
| β-Ionone | [145] | |
| Nerolidol | [145] | |
| OR2C1 | Nonanethiol | [135] |
| Octanethiol | [135,136] | |
| OR2H1 | Methional | [30] |
| OR2H2 | Aldehyde 13-13 | [45] |
| OR2G2 | Cinnamaldehyde | [145] |
| α-Damascone | [145] | |
| Maltyl isobutyrate | [145] | |
| Vanilin | [145] | |
| OR2J2 | Citral | [137] |
| Coumarin | [30,135] | |
| Cyclohexanone | [135] | |
| 1-Decanol | [135] | |
| 2,4-DNT | [137] | |
| Ethyl vanillin | [137] | |
| Eugenol methyl ether | [137] | |
| Eugenyl acetate | [137] | |
| Helional | [137] | |
| 1-Heptanol | [135] | |
| cis-3-Hexen-1-ol | [136] | |
| (+)-Menthol | [137] | |
| Nonanal | [137] | |
| 1-Nonanol | [135] | |
| 1-Octanol | [135,137] | |
| Octanethiol | [137] | |
| Quinoline | [137] | |
| OR2J3 | Cinnamaldehyde | [136] |
| Citral | [137] | |
| Coumarin | [137] | |
| 2,4-DNT | [137] | |
| Eugenol methyl ether | [137] | |
| Geranyl acetate | [148] | |
| Helional | [137] | |
| cis-3-Hexen-1-ol | [136,148] | |
| 1-Octanol | [137] | |
| Musk xylol | [139] | |
| OR2M2 | (-)-β-Citronellol | [134,135] |
| OR2M3 | 3-Mercapto-2-methylpentan-1-ol | [149] |
| OR2M4 | Cinnamaldehyde | [145] |
| Cresyl methyl ether | [145] | |
| α-Damascone | [145] | |
| Estragole | [145] | |
| Fructone | [145] | |
| Nerolidol | [145] | |
| Vanilin | [145] | |
| OR2M7 | (-)-β-Citronellol | [135] |
| Geraniol | [135] | |
| OR2T2 | 1-Pentanethiol | [150] |
| 3-Sulphanyl-1-hexanol | [150] | |
| OR2T4 | Lilial | [151] |
| α-Pinene | [151] | |
| Undecanal | [151] | |
| OR2T8 | 1-Pentanethiol | [150] |
| OR2T10 | Cinnamaldehyde | [145] |
| α-Damascone | [145] | |
| Maltyl isobutyrate | [145] | |
| Terpinyl acetate | [145] | |
| Vanilin | [145] | |
| OR2T11 | Bis (methylthiomethyl) disulfide | [152] |
| 1-Butanethiol | [152] | |
| 2-Butanethiol | [152] | |
| Cyclopentanethiol | [152] | |
| Ethanethiol | [152] | |
| Methanethiol | [152] | |
| 2-Methyl-1-propanethiol | [152] | |
| 3-Methyl-2-butanethiol | [152] | |
| 2-Methyl-2-propanethiol (t-butyl mercaptan; TBM) | [152] | |
| 1-Propanethiol | [152] | |
| 2-Propanethiol | [152] | |
| 2-Pentanethiol | [152] | |
| 2,3,5-Trithiahexane | [152] | |
| Thiolane-2-thiol | [152] | |
| OR2T34 | Cinnamaldehyde | [145] |
| α-Damascone | [145] | |
| Estragole | [145] | |
| Floralozone | [145] | |
| Fructone | [145] | |
| Jasmonyl | [145] | |
| Vanilin | [145] | |
| OR2W1 | Acetophenone | [135] |
| Allyl phenyl acetate | [135,137] | |
| trans-Anethole | [138] | |
| Allylphenyl acetate | [138] | |
| Benzophenone | [135] | |
| Benzyl acetate | [135] | |
| Butyl formate | [135] | |
| Coffee difuran | [137] | |
| Coumarin | [135,137] | |
| (-)-carvone | [135] | |
| (+)-Carvone | [135,137] | |
| (-)-β-Citronellol | [135] | |
| 4-Chromanone | [135] | |
| (R)-(-)-Carvone | [138] | |
| Cinnamyl acetate | [138] | |
| Decanoic acid | [135] | |
| Dihydrojasmone | [135] | |
| (+)-Dihydrocarvone | [135] | |
| 1-Decanol | [135] | |
| p, α-Dimethylstyrene | [138] | |
| Eugenol methyl ether | [137] | |
| Estragole | [138] | |
| Ethyl cyclohexanecarboxylate | [138] | |
| Geraniol | [135] | |
| Helional | [137] | |
| Heptanal | [135] | |
| Hexanal | [135] | |
| Hexyl acetate | [135] | |
| 1-Heptanol | [135] | |
| 1-Hexanol | [135] | |
| 2,3-Hexanedione | [135] | |
| 2-Heptanone | [135] | |
| 2-Hexanone | [135] | |
| cis-3-Hexen-1-ol | [136] | |
| 3,4-Hexanedione | [135] | |
| 3-Heptanone | [135] | |
| (E)-2-Heptenal | [138] | |
| 2-Heptanol | [138] | |
| Hexyl acetate | [138] | |
| (S)-(-)-Limonene | [138] | |
| (R)-(+)-Limonene | [137,138] | |
| 2-Methyl-2-propanethiol (t-butyl mercaptan; TBM) | [152] | |
| Methyl salicylate | [137] | |
| 3-Mercaptohexyl acetate | [138] | |
| Methyl cinnamate | [138] | |
| 3-Methylbutylacetate | [138] | |
| 4-Methyl acetophenone | [138] | |
| Nonanethiol | [135] | |
| Nonanoic acid | [135,137] | |
| Nonanal | [135,137] | |
| 1-Nonanol | [135] | |
| 2-Nonanone | [135] | |
| Nonanal | [138] | |
| 2-Nonanone | [138] | |
| 2-Octanone | [135] | |
| Octanoic acid | [135] | |
| 3-Octanone | [135] | |
| Octanethiol | [135,137] | |
| 1-Octanol | [135,137] | |
| Octanal | [138] | |
| (R/S)-Octen-3-ol | [138] | |
| (R/S)-γ-Octalactone | [138] | |
| Prenyl acetate | [135] | |
| 2-Pentylpyridine | [138] | |
| 2-Phenylethyl acetate | [138] | |
| 2-Phenylethanethiol | [138] | |
| OR2W3 | Nerol | [30] |
| OR2Y1 | (-)-Carvone | [134] |
| OR3A1 | Aldehyde TPM | [153] |
| Bourgeonal (Antagonist) | [153] | |
| Cyclosal | [153] | |
| Foliaver | [153] | |
| Helional | [153,154] | |
| Heliotropyl acetone | [154] | |
| Hydrocinnamaldehyde (Antagonist) | [153] | |
| Lilial | [153] | |
| Methyl-hydrocinnamaldehyde | [153] | |
| Methyl-phenyl-pentanal | [153] | |
| Methylcinnamaldehyde (Antagonist) | [153] | |
| Trifernal | [153] | |
| OR4D1 | PI-23472 | [30,42] |
| OR4D6 | β-Ionone | [85] |
| OR4D9 | β-Ionone | [85] |
| OR4E2 | Amyl acetate | [136] |
| OR4M1 | Acetophenone | [155] |
| OR5A1 | β-Ionone | [85] |
| OR5A2 | β-Ionone | [85] |
| OR5AC2 | α-Damascone | [145] |
| Eugenyl acetate | [145] | |
| Fructone | [145] | |
| Maltyl isobutyrate | [145] | |
| Manzanate | [145] | |
| Vanilin | [145] | |
| OR5AN1 | Ambretone | [139,141] |
| Ambrettolide | [139] | |
| Cosmone | [139] | |
| Cyclopentadecanone | [139,141] | |
| Cyclopentadecanol | [141] | |
| Civetone | [141] | |
| Dihydrocivetone | [141] | |
| (R)-6,6-Difluoromuscone | [141] | |
| (R)-7,7-Difluoromuscone | [141] | |
| (R)-8,8-Difluoromuscone | [141] | |
| (R)-10,10-Difluoromuscone | [141] | |
| (R)-6,7-Dehydromuscone (E:Z = 10:1) | [141] | |
| (R,Z)-6,7-Dehydromuscone | [141] | |
| (R)-9,10-Dehydro-6,6-difluoromuscone (E:Z = 3:2) | [141] | |
| (R,E)-10,11-Dehydro-7,7-dilfuoromuscone | [141] | |
| (R,Z)-10,11-Dehydro-7,7-dilfuoromuscone | [141] | |
| (R)-5,6-Dehydro-8,8-difluoromuscone (E:Z = 10:1) | [141] | |
| (R,E)-6,7-Dehydro-9,9-difluoromuscone | [141] | |
| (R,E)-6,7-Dehydro-10,10-difluoromuscone | [141] | |
| (R,Z)-6,7-Dehydro-10,10-difluoromuscone | [141] | |
| Exaltolide | [139] | |
| Ethylenebrassylate | [139] | |
| Globanone | [139] | |
| 3-Methylcyclotetradecanone | [139] | |
| Habanolide | [139] | |
| Isomuscone | [141] | |
| Muscone | [139,156] | |
| Musk xylol | [139,141] | |
| Musk ketone | [139] | |
| racemic-Muscone | [139,141] | |
| l-Muscone | [139] | |
| d-Muscone | [139] | |
| Musk ketone | [139,141] | |
| Musk tibetene | [141] | |
| (R)-Muscone | [141] | |
| Muscenone | [139] | |
| ω-Pentadecalactone | [141] | |
| Thiacyclopentadecane 1-oxide | [141] | |
| OR5B17 | Eugenyl acetate | [145] |
| Floralozone | [145] | |
| OR5D3P | Raspberry ketone | [157] |
| OR5D18 | 2-Acetyl-3,5(6)-dimethylpyrazine (mixture of isomers) | [158] |
| 2-Acetyl-3-ethylpyrazine | [158] | |
| 2-Acetyl-3-methylpyrazine | [158] | |
| Benzene | [137] | |
| Eugenol | [63] | |
| OR5K1 | Citral | [137] |
| β-Damascone | [137] | |
| 2,3-Diethyl-5-methylpyrazine | [158] | |
| 2,3-Diethylpyrazine | [158] | |
| 2,3-Dimethylpyrazine | [158] | |
| 2,5-Dihydro-2,4,5-trimethylthiazoline | [158] | |
| 2,5-Dimethylpyrazine | [158] | |
| 2,5-Dimethylpyrazine | [158] | |
| 2-Ethyl-3,5(6)-dimethylpyrazine | [158] | |
| 2-Ethyl-3-methoxypyrazine | [158] | |
| 2-Ethyl-3-methylpyrazine | [158] | |
| 2-Ethyl-5(6)-methylpyrazine, mixture of isomers | [158] | |
| Eugenol | [137] | |
| Eugenol methyl ether | [137] | |
| 2-Heptanone | [137] | |
| 2-Isobutyl-3-methoxypyrazine | [158] | |
| 2-Isopropyl-3-methoxypyrazine | [158] | |
| (R)-(+)-Limonene | [137] | |
| Lyral | [137] | |
| 2-Methoxypyrazine | [158] | |
| (2R/2S)-4-Methoxy-2,5-dimethylfuran-3(2H)-one, sum of isomers (Methoxyfuraneol) | [158] | |
| 5H-5-Methyl-6,7-dihydrocyclopenta-[b] pyrazine | [158] | |
| Nonanal | [137] | |
| Propanal | [137] | |
| Quinine | [137] | |
| 2,3,5-Trimethylpyrazine | [158] | |
| 2-Vinylpyrazine | [158] | |
| OR5P3 | Acetophenone | [135] |
| Coumarin | [135,137] | |
| (-)-Carvone | [135] | |
| (+)-Carvone | [135,137] | |
| 1-Heptanol | [135] | |
| 1-Hexanol | [135] | |
| Quinoline | [137] | |
| OR6P1 | Anisaldehyde | [136] |
| OR6X1 | (-)-Carvone | [134] |
| OR7A5 | Myrac | [30,42] |
| OR7C1 | Androstadienone | [136] |
| OR7D4 | Androstadienone | [159] |
| Androstenone | [159] | |
| OR8B3 | (+)-Carvone | [136] |
| OR8D1 | Caramel furanone | [136] |
| Sotolone | [30] | |
| OR8H1 | Scatole | [160] |
| OR8K3 | β-Damascenone | [137] |
| (+)-Menthol | [136,137] | |
| OR10A6 | Benzyl acetone | [83] |
| α-Cinnamyl alcohol | [83] | |
| Citronellol | [83] | |
| Cyclamen aldehyde | [83] | |
| Cyclemone A | [83] | |
| Geraniol | [83] | |
| α-Ionone | [83] | |
| Nerol | [83] | |
| Nonadecane | [83] | |
| Linalool | [83] | |
| Lyral | [83] | |
| Phenyl ethyl alcohol | [83] | |
| Phenyl propyl alcohol | [83] | |
| 3-Phenyl propyl propionate | [83,136,161] | |
| OR10G3 | Ethyl vanillin | [136,137] |
| Eugenol | [137] | |
| Vanillin | [136] | |
| OR10G4 | Ethyl vanillin | [137] |
| Vanillin | [136] | |
| OR10G7 | Ethyl vanillin | [137] |
| Eugenol | [136,137] | |
| Eugenyl acetate | [137] | |
| Eugenol methyl ether | [137] | |
| OR10G9 | Ethyl vanillin | [137] |
| OR10H1 | Sandranol | [119] |
| OR10J1 | Dimetol | [30] |
| OR10J5 | Lyral | [135,136,137] |
| α-Cedrene | [46] | |
| OR10S1 | Lilial | [151] |
| OR11A1 | 2-Ethyl fenchol | [136,137] |
| OR11H4 | Isovaleric acid | [162] |
| Phenyl ethyl alcohol | [83] | |
| Phenyl propyl alcohol | [83] | |
| Skatole | [163] | |
| OR11H6 | Isovaleric acid | [162] |
| OR11H7P | Isovaleric acid | [162] |
| OR51B4 | Troenan | [117] |
| OR51B5 | Isononyl alcohol | [76] |
| OR51E1 | Butyric acid | [137,164] |
| Butyl butyryllactate | [135] | |
| Dodecanoic acid | [164] | |
| Decanoic acid | [164] | |
| Dimethyl disulfide | [137] | |
| 2,4-DNT | [137] | |
| Eugenol methyl ether | [137] | |
| Eugenyl acetate | [137] | |
| 2-Ethylhexanoic acid (Antagonist) | [164] | |
| Heptanoic acid | [164] | |
| Hexanoic acid | [164] | |
| Isovaleric acid | [136] | |
| Methyl furfuryl disulfide | [137] | |
| Methyl salicylate | [137] | |
| (+)-Menthol | [137] | |
| 3-Methyl-valeric acid | [165] | |
| 4-Methyl-valeric acid | [165] | |
| 4-Methylnonanoic acid | [164] | |
| Nonanoic acid | [30,135] | |
| 2-Nonenoic acid | [164] | |
| Octanoic acid | [164] | |
| 1-Pentanol | [137] | |
| Propanal | [137] | |
| Pyrazine | [137] | |
| Pentanoic acid | [164] | |
| Tetradecanoic acid | [164] | |
| Trans-2-decenoic acid | [164] | |
| Tridecanoic acid | [164] | |
| Undecanoic acid | [164] | |
| OR51E2 | Acetate | [7,90] |
| AFMK | [113] | |
| Androstanedione | [113] | |
| Adenosine-2′,3′-c-phosphate | [113] | |
| D-Alanyl-d-alanine | [113] | |
| N-Acetylglutamic acid | [113] | |
| Bradykinin | [113] | |
| Epitestosterone | [113] | |
| Estriol | [113] | |
| Glycine | [113] | |
| γ-CEHC | [113] | |
| L-Glyceric acid | [113] | |
| Hydroxypyruvic acid | [113] | |
| L-Histidinol | [113] | |
| 8-Hydroxyguanine | [113] | |
| β-Ionone | [19,30] | |
| α-Ionone (Antagonist) | [19] | |
| α-Ionone (Agonist) | [166] | |
| Imidazolone | [113] | |
| Kojibiose | [113] | |
| 2-Ketoglutaric acid | [113] | |
| 19-OH AD | [113] | |
| Propionic acid | [7,135] | |
| 2-Pyrrolidinone | [113] | |
| Pelargonidin | [113] | |
| Palmitic acid | [113] | |
| Tetrahydrocurcumin | [113] | |
| Urea | [113] | |
| OR51I2 | Isovaleric acid | [167] |
| OR51L1 | Allyl phenyl acetate | [135,136,137] |
| Hexanoic acid | [135] | |
| OR52A5 | 4-Ethyloctanoic acid | [168] |
| OR52B2 | Decanoic acid | [168] |
| OR52D1 | Acetophenone | [143] |
| Anisole | [143] | |
| Benzaldehyde | [143] | |
| Benzothiazol | [143] | |
| β-Ionone | [143] | |
| Butyl butyrate | [143] | |
| Butyric acid | [143] | |
| Caproic acid | [143] | |
| Caprylic acid | [143] | |
| Cinnamaldehyde | [143] | |
| Citral | [143] | |
| Citralva | [143] | |
| Cyclohexanone | [143] | |
| Decanal | [143] | |
| 2-Decanone | [143] | |
| Estragol | [143] | |
| Ethyl butyrate | [143] | |
| Ethyl caproate | [143] | |
| Ethyl heptanoate | [143] | |
| Helional | [143] | |
| Heptanoic acid | [143] | |
| 3-Hydroxybutan-2-one | [143] | |
| Isoamyl acetate | [143] | |
| Isobutyric acid | [143] | |
| Isovaleric acid | [143] | |
| 2-Isobutyl-3-methoxypyrazine | [143] | |
| Lauric aldehyde | [143] | |
| Methyl heptanoate | [143] | |
| Methyl octanoate | [30,143] | |
| 6-Methyl-5-hepten-2-one | [143] | |
| Nonanal | [143] | |
| Nonanoic acid | [143] | |
| 3-Nonanone | [143] | |
| 2-Nonanol | [143] | |
| 1-Nonanol | [143] | |
| Octanal | [143] | |
| 3-Octanone | [143] | |
| Octanic acid | [134,143] | |
| Octanic acid methyl ester | [134,143] | |
| Para-anisaldehyde | [143] | |
| Phenylmethanol | [143] | |
| Propionic acid | [143] | |
| Safrole | [143] | |
| S-methylthio butanoate | [143] | |
| Thiazol | [143] | |
| Trans-anethol | [143] | |
| Trans-cinnamic acid | [143] | |
| OR52E1 | Butanoic acid | [168] |
| OR52E8 | 3-Hydroxy-3-methylhexanoic acid | [168] |
| OR52L1 | Pentanoic acid | [168] |
| OR56A1 | Decanal | [137] |
| OR56A4 | Decanal | [137] |
| Undecanal | [136] | |
| OR56A5 | Decanal | [137] |
| OR56B4 | δ-Decalactone | [16] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tong, T.; Wang, Y.; Kang, S.-G.; Huang, K. Ectopic Odorant Receptor Responding to Flavor Compounds: Versatile Roles in Health and Disease. Pharmaceutics 2021, 13, 1314. https://0-doi-org.brum.beds.ac.uk/10.3390/pharmaceutics13081314
Tong T, Wang Y, Kang S-G, Huang K. Ectopic Odorant Receptor Responding to Flavor Compounds: Versatile Roles in Health and Disease. Pharmaceutics. 2021; 13(8):1314. https://0-doi-org.brum.beds.ac.uk/10.3390/pharmaceutics13081314
Chicago/Turabian StyleTong, Tao, Yanan Wang, Seong-Gook Kang, and Kunlun Huang. 2021. "Ectopic Odorant Receptor Responding to Flavor Compounds: Versatile Roles in Health and Disease" Pharmaceutics 13, no. 8: 1314. https://0-doi-org.brum.beds.ac.uk/10.3390/pharmaceutics13081314