In-Field Detection of Yellow Rust in Wheat on the Ground Canopy and UAV Scale
Abstract
:1. Introduction
2. Materials and Methods
2.1. Field Trial Layout
2.1.1. Inoculations
2.1.2. Visual Disease Ratings
2.1.3. Crop Stand and Disease Development
2.2. Measurement Platforms
2.2.1. Field Platform Phytobike
2.2.2. UAV Measurements
2.3. Data Preprocessing
2.3.1. Spectral Preprocessing
2.3.2. Data Normalization
2.4. Prediction Algorithms
2.4.1. Spectral Angle Mapper
2.4.2. Support Vector Algorithms
2.5. Vegetation Indices
2.6. Model Evaluation
2.7. Feature Selection
2.8. Spatial Resolution as a Key Parameter for Disease Detection
3. Results and Discussion
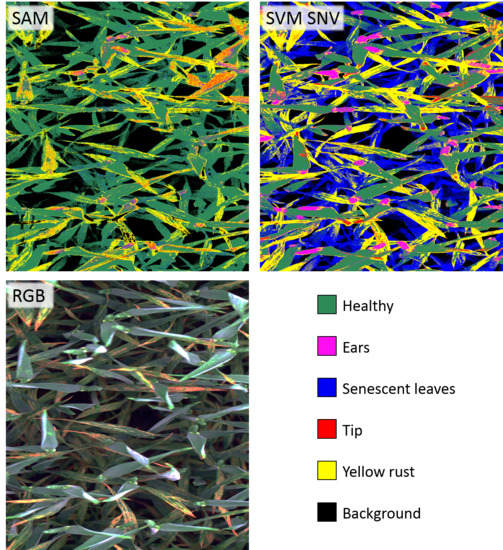
3.1. Supervised Classification of Hyperspectral Pixels at the Ground Canopy Scale
3.2. Evaluation of Hyperspectral UAV Observations Using a Filter-System Hyperspectral Camera
3.3. Selection of Relevant Features at Different Scales
3.3.2. Ground Scale
3.3.3. UAV Scale
3.3.4. Cross-Scale Interpretation
3.3.5. Spatial Resolution as Key Parameter for Disease Detection
3.4. Optimal Sensor System for Plant Disease Detection
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- West, J.; Bravo, C.; Oberti, R.; Lemaire, D.; Moshou, D.; McCartney, A. The potential of optical canopy measurement for targeted control of field crop diseases. Ann. Rev. Phytopathol. 2003, 41, 593–614. [Google Scholar] [CrossRef] [PubMed]
- Hillnhütter, C.; Mahlein, A.K.; Sikora, R.A.; Oerke, E.C. Remote sensing to detect plant stress induced by Heterodera schachtii and Rhizoctonia solani in sugar beet fields. Field Crops Res. 2011, 122, 70–77. [Google Scholar] [CrossRef]
- Bravo, C.; Moshou, D.; West, J.; McCartney, A.; Ramon, H. Early disease detection in wheat fields using spectral reflectance. Biosyst. Eng. 2003, 84, 137–145. [Google Scholar] [CrossRef]
- Mewes, T.; Franke, J.; Menz, G. Spectral requirements on airborne hyperspectral remote sensing data for wheat disease detection. Precis. Agric. 2011, 12, 795–812. [Google Scholar] [CrossRef]
- Mirik, M.; Jones, D.C.; Price, J.A.; Workneh, F.; Ansley, R.J.; Rush, C.M. Satellite remote sensing of wheat infected by wheat streak mosaic virus. Plant Dis. 2011, 95, 4–12. [Google Scholar] [CrossRef]
- Gebbers, R.; Adamchuk, V.I. Precision agriculture and food security. Science 2010, 327, 828–831. [Google Scholar] [CrossRef]
- Mahlein, A.K.; Kuska, M.T.; Behmann, J.; Polder, G.; Walter, A. Hyperspectral sensors and imaging technologies in phytopathology: State of the art. Ann. Rev. Phytopathol. 2018, 56, 535–558. [Google Scholar] [CrossRef]
- Wahabzada, M.; Mahlein, A.K.; Bauckhage, C.; Steiner, U.; Oerke, E.C.; Kersting, K. Plant phenotyping using probabilistic topic models: Uncovering the hyperspectral language of plants. Sci. Rep. 2016, 6, 22482. [Google Scholar] [CrossRef]
- Whetton, R.; Hassall, K.; Waine, T.W.; Mouazen, A. Hyperspectral measurements of yellow rust and fusarium head blight in cereal crops: Part 1: Laboratory study. Biosyst. Eng. 2017, 166, 101–115. [Google Scholar] [CrossRef]
- Whetton, R.; Waine, T.; Mouazen, A. Hyperspectral measurements of yellow rust and fusarium head blight in cereal crops: Part 2: On-line field measurement. Biosyst. Eng. 2018, 167, 144–158. [Google Scholar] [CrossRef] [Green Version]
- Mahlein, A.K. Plant disease detection by imaging sensors—Parallels and scientific demands for precision agriculture and plant phenotyping. Plant Dis. 2016, 100, 241–251. [Google Scholar] [CrossRef] [PubMed]
- Thomas, S.; Kuska, M.T.; Bohnenkamp, D.; Brugger, A.; Alisaac, E.; Wahabzada, M.; Behmann, J.; Mahlein, A.K. Benefits of hyperspectral imaging for plant disease detection and plant protection: A technical perspective. J. Plant Dis. Prot. 2018, 125, 5–20. [Google Scholar] [CrossRef]
- Bravo, C.; Moshou, D.; Oberti, R.; West, J.; McCartney, A.; Bodria, L.; Ramon, H. Foliar disease detection in the field using optical sensor fusion. Agric. Eng. Int. 2004, 6. [Google Scholar]
- Franke, J.; Menz, G. Multi-temporal wheat disease detection by multi-spectral remote sensing. Precis. Agric. 2007, 8, 161–172. [Google Scholar] [CrossRef]
- Huang, W.; Lamb, D.W.; Niu, Z.; Zhang, Y.; Liu, L.; Wang, J. Identification of yellow rust in wheat using in-situ spectral reflectance measurements and airborne hyperspectral imaging. Precis. Agric. 2007, 8, 187–197. [Google Scholar] [CrossRef]
- Lan, Y.B.; Chen, S.D.; Fritz, B.K. Current status and future trends of precision agricultural aviation technologies. Int. J. Agric. Biol. Eng. 2017, 10, 1–17. [Google Scholar]
- Devadas, R.; Lamb, D.W.; Simpfendorfer, S.; Backhouse, D. Evaluating ten spectral vegetation indices for indentifying rust infection in individual wheat leaves. Precis. Agric. 2009, 10, 459–470. [Google Scholar] [CrossRef]
- Ashourloo, D.; Mobasheri, M.R.; Huete, A. Developing two spectral indices for detection of wheat leaf rust (Puccinia triticina). Remote Sens. 2014, 6, 4723–4740. [Google Scholar] [CrossRef]
- Cao, X.; Luo, Y.; Zhou, Y.; Fan, J.; Xu, X.; West, J.S.; Duan, X.; Cheng, D. Detection of powdery mildew in two winter wheat plant densities and prediction of grain yield using canopy hyperspectral reflectance. PLoS ONE 2015, 10. [Google Scholar] [CrossRef]
- Blackburn, G.A. Hyperspectral remote sensing of plant pigments. J. Exp. Bot. 2007, 58, 855–867. [Google Scholar] [CrossRef]
- Gitelson, A.A.; Keydan, G.P.; Merzlyak, M.N. Three-band model for noninvasive estimation of chlorophyll, carotenoids, and anthocyanin contents in higher plant leaves. Geophys. Res. Lett. 2006, 33, L11402. [Google Scholar] [CrossRef]
- Roosjen, P.P.J.; Suomalainen, J.M.; Bartholomeus, H.M.; Clevers, J.G.P.W. Hyperspectral reflectance anisotropy measurements using a pushbroom spectrometer on an unmanned aerial vehicle—Results for barley, winter wheat and potato. Remote Sens. 2016, 8, 909. [Google Scholar] [CrossRef]
- Moshou, D.; Bravo, C.; West, J.; Wahlen, T.; McCartney, A.; Ramon, H. Automatic detection of ‘yellow rust’ in wheat using reflectance measurements and neural networks. Comput. Electron. Agric. 2004, 44, 173–188. [Google Scholar] [CrossRef]
- Zhang, J.; Pu, R.; Huang, W.J.; Yuan, L.; Luo, J.; Wang, J. Using in-situ hyperspectral data for detecting and discriminating yellow rust disease from nutrient stresses. Field Crop Res. 2012, 134, 165–174. [Google Scholar] [CrossRef]
- Zheng, Q.; Huang, W.; Cui, X.; Dong, Y.; Shi, Y.; Ma, H.; Liu, L. Identification of wheat yellow rust using optimal three-band spectral indices in different growth stages. Sensors 2019, 19, 35. [Google Scholar] [CrossRef]
- Azadbakht, M.; Ashourloo, D.; Aghighi, H.; Radiom, S.; Alimohammadi, A. Wheat leaf rust detection at canopy scale under different LAI levels using machine learning techniques. Comput. Electron. Agric. 2019, 156, 119–128. [Google Scholar] [CrossRef]
- Chen, W.; Wellings, C.; Chen, X.; Kang, Z.; Liu, T. Wheat stripe (yellow) rust caused by Puccinia striiformis f. sp. tritici. Mol. Plant. Pathol. 2014, 15, 433–446. [Google Scholar] [CrossRef]
- Yu, K.; Anderegg, J.; Mikaberidze, A.; Petteri, K.; Mascher, F.; McDonald, B.A.; Walter, A.; Hund, A. Hyperspectral canopy sensing of wheat Septoria tritici blotch disease. Front. Plant Sci. 2018, 9, 1195. [Google Scholar] [CrossRef]
- Behmann, J.; Mahlein, A.K.; Rumpf, T.; Romer, C.; Plümer, L. A review of advanced machine learning methods for the detection of biotic stress in precision crop protection. Precis. Agric. 2015, 16, 239–260. [Google Scholar] [CrossRef]
- Camps-Valls, G.; Tuia, D.; Bruzzone, L.; Benediktsson, J.A. Advances in hyperspectral image classification: Earth monitoring with statistical learning methods. IEEE Signal Process. Mag. 2014, 31, 45–54. [Google Scholar] [CrossRef]
- Lowe, A.; Harrison, N.; French, A.P. Hyperspectral image analysis techniques for the detection and classification of the early onset of plant disease and stress. Plant Methods 2017, 13, 80. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Ganapathysubramanian, B.; Singh, A.K.; Sarkar, S. Machine learning for high-throughput stress phenotyping in plants. Trends Plant Sci. 2016, 21, 110–124. [Google Scholar] [CrossRef] [PubMed]
- Barnes, R.J.; Dhanoa, M.S.; Lister, S.J. Standard normal variate transformation and de-trending of near-infrared diffuse reflectance spectra. Appl. Spectrosc. 1989, 43, 772–777. [Google Scholar] [CrossRef]
- Kruse, F.A.; Lefkoff, A.B.; Boardman, J.W.; Heidebrecht, K.B.; Shapiro, A.T.; Barloon, P.J.; Goetz, A.F.H. The spectral image processing system (SIPS)—Interactive visualization and analysis of imaging spectrometer data. Remote Sens. Environ. 1993, 44, 145–163. [Google Scholar] [CrossRef]
- Cortes, C.; Vapnik, V. Support-Vector networks. Mach. Learn. 1995, 20, 273–297. [Google Scholar] [CrossRef]
- Chang, C.C.; Lin, C.J. LIBSVM: A library for support vector machines. ACM Trans. Intell. Syst. Technol. 2011, 2, 27. [Google Scholar] [CrossRef]
- Behmann, J.; Steinrücken, J.; Plümer, L. Detection of early plant stress responses in hyperspectral images. ISPRS 2014, 93, 98–111. [Google Scholar] [CrossRef]
- Guyon, I.; Elisseeff, A. An introduction to variable and feature selection. J. Mach. Learn. Res. 2003, 3, 1157–1182. [Google Scholar]
- Parker, S.R.; Shaw, M.W.; Royle, D.J. The reliability of visual estimates of disease severity on cereal leaves. Plant Pathol. 1995, 44, 856–864. [Google Scholar] [CrossRef]
- Nutter, F.W.; Gleason, M.L.; Jenco, J.H.; Christians, N.C. Assessing the accuracy, intra-rater repeatability, and inter-rater reliability of disease assessment systems. Phytopathology 1993, 83, 806–812. [Google Scholar] [CrossRef]
- Mahlein, A.K.; Rumpf, T.; Welke, P.; Dehne, H.W.; Plümer, L.; Steiner, U.; Oerke, E.C. Development of spectral indices for detectiong and identifying plant diseases. Remote Sens. Environ. 2013, 128, 21–30. [Google Scholar] [CrossRef]
- Chen, T.; Zhang, J.; Chen, Y.; Wan, S.; Zhang, L. Detection of peanut leaf spots disease using canopy hyperspectral reflectance. Comput. Electron. Agric. 2019, 156, 677–683. [Google Scholar] [CrossRef]
- Gates, D.M.; Keegan, H.J.; Schelter, J.C.; Weidner, V.R. Spectral properties of plants. Appl. Opt. 1965, 4, 11–20. [Google Scholar] [CrossRef]
- Curran, P.J. Remote sensing of foliar chemistry. Remote Sens. Environ. 1989, 30, 271–278. [Google Scholar] [CrossRef]
- Heim, R.H.J.; Jurgens, N.; Große-Stoltenberg, A.; Oldeland, J. ; The effect of epidermal structures on leaf spectral signatures of ice plants (Aizoaceae). Remote Sens. 2015, 7, 16901–16914. [Google Scholar] [CrossRef]
- Carter, G.A.; Knapp, A.K. Leaf optical properties in higher plants: Linking spectral characteristics to stress and chlorophyll concentrations. Am. J. Bot. 2001, 88, 677–684. [Google Scholar] [CrossRef]
- Pandey, P.; Ge, Y.; Stoerger, V.; Schnable, J.C. High throughput in vivo analysis of plant leaf chemical properties using hyperspectral imaging. Front. Plant Sci. 2017, 8, 1348. [Google Scholar] [CrossRef]
- Gay, A.; Thomas, H.; James, C.; Taylor, J.; Rowland, J.; Ougham, H. Nondestructive analysis of senescence in mesophyll cells by spectral resolution of protein synthesis-dependent pigment metabolism. New Phytol. 2008, 179, 663–674. [Google Scholar] [CrossRef]
- Bohnenkamp, D.; Kuska, M.T.; Mahlein, A.K.; Behmann, J. Hyperspectral signal decomposition and symptom detection of wheat rust disease at the leaf scale using pure fungal spore spectra as reference. Plant Pathol. 2019, 68, 1188–1195. [Google Scholar] [CrossRef]
- Elvidge, C.D. Visible and near infrared reflectance characteristics of dry plant materials. Int. J. Remote Sens. 1990, 11, 1775–1795. [Google Scholar] [CrossRef]
- Kuska, M.; Wahabzada, M.; Leucker, M.; Dehne, H.W.; Kersting, K.; Oerke, E.C.; Steiner, U.; Mahlein, A.K. Hyperspectral phenotyping on the microscopic scale: Towards automated characterization of plant-pathogen interactions. Plant Methods 2015, 11, 28. [Google Scholar] [CrossRef] [PubMed]









| SVM Raw | SVM SNV | SVM Indices | SAM | |
|---|---|---|---|---|
| Accuracy | 91.9% | 92.9% | 90.2% | 81.4% |
| F1 disease score | 83.2% | 84.0% | 76.4% | 48.3% |
| Trait/Treatment | Performance |
|---|---|
| Fertilizer level | Accuracy = 82.3% |
| Fungicide | Accuracy = 91.5% |
| Fungicide + fertilizer level (four classes) | Accuracy = 71.4% |
| Disease detection (severity > 0) | Accuracy = 90.0% |
| Disease severity estimation | Correlation = 70.6% |
| Ranking | Fert | Fung | Fert + Fung | YR Detection | YR Regression |
|---|---|---|---|---|---|
| 1 | 767 | 727 | 734 | 797 | 832 |
| 2 | 725 | 804 | 887 | 881 | 510 |
| 3 | 648 | 762 | 545 | 601 | 867 |
| 4 | 557 | 648 | 559 | 706 | 874 |
| 5 | 627 | 594 | 517 | 874 | 587 |
| 6 | 704 | 767 | 748 | 594 | 594 |
| Ground Class. | All | UAV Select | Field Select | Equidistant | VI |
|---|---|---|---|---|---|
| # feature | 210 | 10 | 10 | 10 | 16 |
| Acc. | 92.9% | 87.4% | 88.9% | 89.2% | 90.2% |
| F1 disease | 0.84 | 0.694 | 0.751 | 0.732 | 0.764 |
| UAV Regression | All | UAV Select | Field Select | Equidistant |
|---|---|---|---|---|
| # feature | 55 | 10 | 10 | 10 |
| R² | 0.63 | 0.69 | 0.57 | 0.61 |
| Corr. | 79.4% | 83.0% | 75.5% | 78.1% |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bohnenkamp, D.; Behmann, J.; Mahlein, A.-K. In-Field Detection of Yellow Rust in Wheat on the Ground Canopy and UAV Scale. Remote Sens. 2019, 11, 2495. https://0-doi-org.brum.beds.ac.uk/10.3390/rs11212495
Bohnenkamp D, Behmann J, Mahlein A-K. In-Field Detection of Yellow Rust in Wheat on the Ground Canopy and UAV Scale. Remote Sensing. 2019; 11(21):2495. https://0-doi-org.brum.beds.ac.uk/10.3390/rs11212495
Chicago/Turabian StyleBohnenkamp, David, Jan Behmann, and Anne-Katrin Mahlein. 2019. "In-Field Detection of Yellow Rust in Wheat on the Ground Canopy and UAV Scale" Remote Sensing 11, no. 21: 2495. https://0-doi-org.brum.beds.ac.uk/10.3390/rs11212495