A Multiscale Hierarchical Model for Sparse Hyperspectral Unmixing
Abstract
:1. Introduction
2. Theoretical Methodology
2.1. Linear Mixture Model
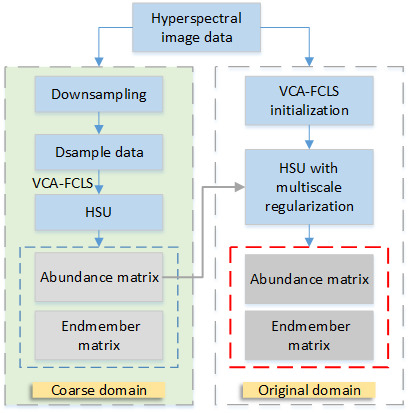
2.2. Multiscale Hierarchical Unmixing Methods
2.2.1. Multiscale Methods
2.2.2. The Optimization Problem
3. Experiments
3.1. Experimental Data
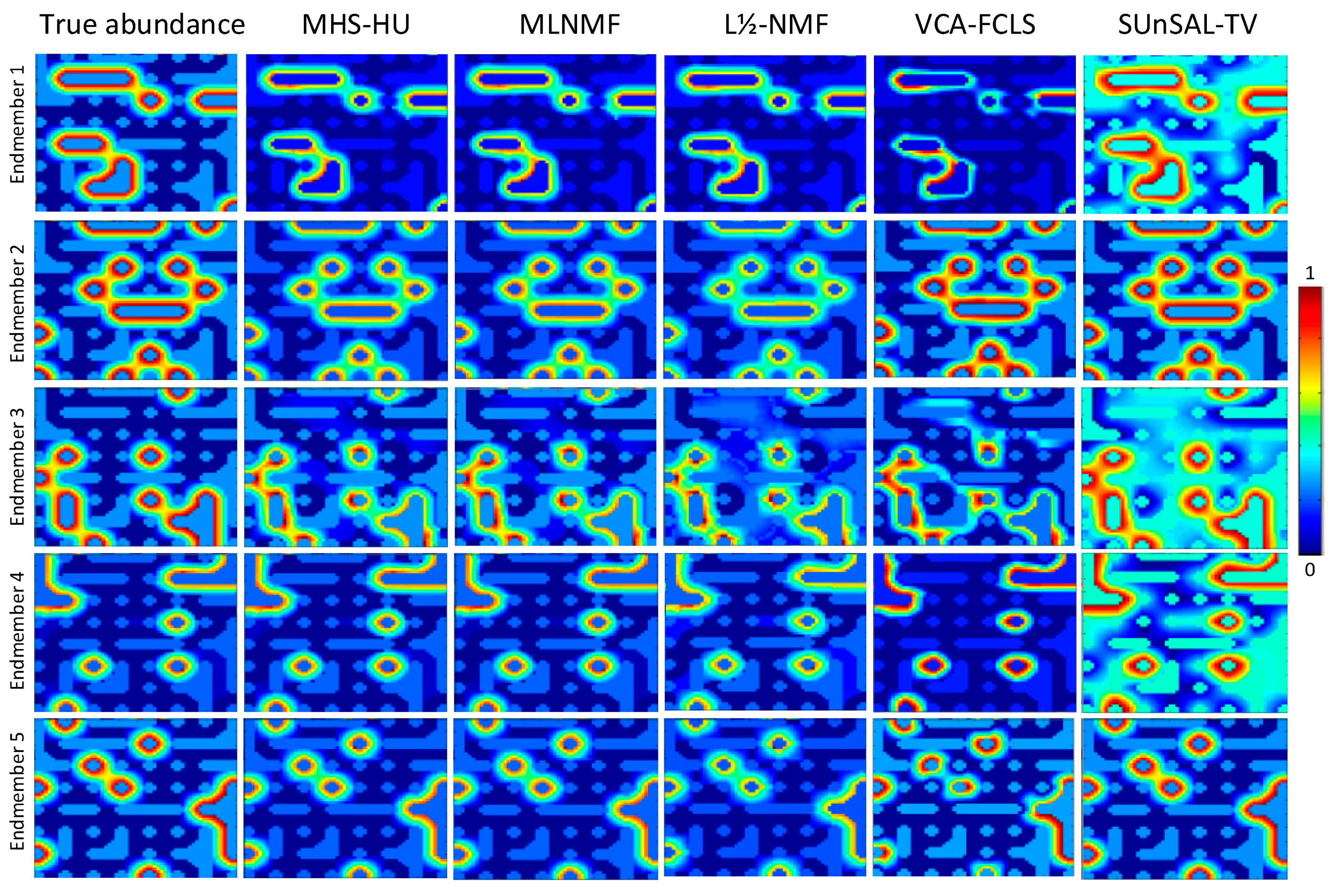
3.2. Simulated experiments
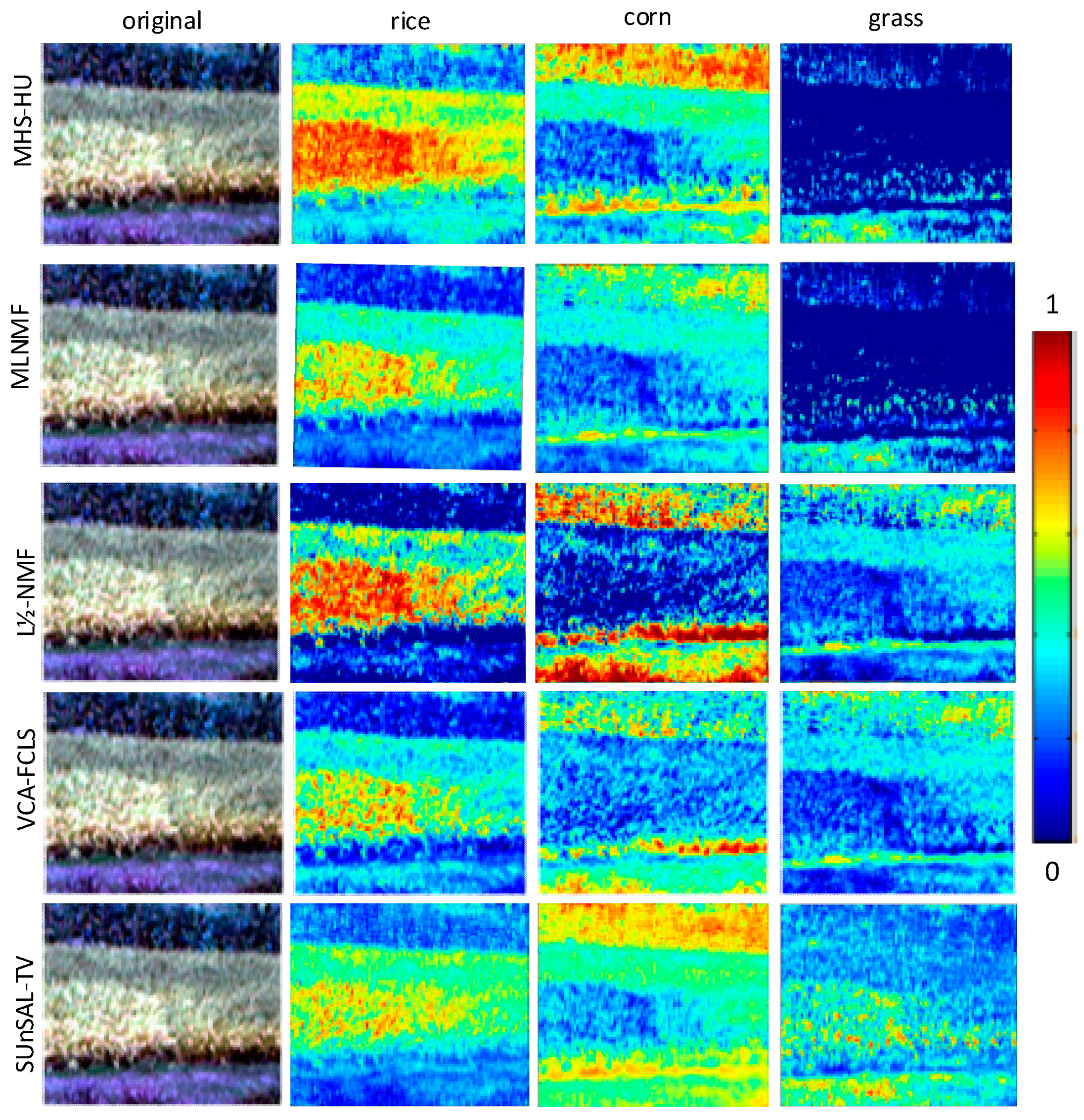
3.3. Real Data Experiments
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Zhang, Y.; Wu, K.; Du, B.; Zhang, L.; Hu, X. Hyperspectral target detection via adaptive joint sparse representation and multi-task learning with locality information. Remote Sens. 2017, 9, 482. [Google Scholar] [CrossRef]
- He, Z.; Wang, Y.; Hu, J. Joint sparse and low-rank multitask learning with laplacian-like regularization for hyperspectral classification. Remote Sens. 2018, 10, 322. [Google Scholar] [CrossRef]
- Bioucas-Dias, J.M.; Plaza, A.; Camps-Valls, G.; Scheunders, P.; Nasrabadi, N.; Chanussot, J. Hyperspectral remote sensing data analysis and future challenge. IEEE Geosci. Remote Sens. Mag. 2013, 1, 6–36. [Google Scholar] [CrossRef]
- Gevaert, C.; Suomalainen, J.; Tang, J.; Kooistra, L. Generation of spectral-temporal response surfaces by combining multispectral satellite and hyperspectral uav imagery for precision agriculture applications. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2015, 8, 3140–3146. [Google Scholar] [CrossRef]
- Ma, D.; Yuan, Y.; Wang, Q. Hyperspectral Anomaly Detection via Discriminative Feature Learning with Multiple-Dictionary Sparse Representation. Remote Sens. 2018, 10, 745. [Google Scholar] [CrossRef]
- Huang, R.; Li, X.; Zhao, L. Hyperspectral unmixing based on incremental kernel nonnegative matrix factorization. IEEE Trans. Geosci. Remote Sens. 2018, 11, 6645–6662. [Google Scholar] [CrossRef]
- Hoyer, P.O. Non-negative matrix factorization with sparseness constraints. J. Mach. Learn. Res. 2004, 5, 1457–1469. [Google Scholar]
- Qian, Y.; Jia, S.; Zhou, J.; Robles-Kelly, A. Hyperspectral unmixing via L1/2 sparsity-constrained nonnegative matrix factorization. IEEE Trans. Geosci. Remote Sens. 2014, 49, 4282–4297. [Google Scholar] [CrossRef]
- Lee, D.D.; Seung, H.S. Algorithms for non-negative matrix factorization. In Proceedings of the International Conference on Neural Information Processing Systems, Denver, CO, USA, 29 November–4 December 2000; MIT Press: Cambridge, MA, USA, 2000. [Google Scholar]
- Ma, W.; Bioucas-Dias, J.; Chan, T.; Gillis, N. A signal processing perspective on hyperspectral unmixing: Insights from remote sensing. IEEE Signal Process. Mag. 2014, 31, 67–81. [Google Scholar] [CrossRef]
- Fang, H.; Li, H.C.; Li, J.; Du, Q.; Plaza, A.; Emery, W.J. Sparsity-constrained deep nonnegative matrix factorization for hyperspectral unmixing. IEEE Trans. Geosci. Remote Sens. 2018, 10, 6245–6257. [Google Scholar] [CrossRef]
- Wang, M.; Zhang, B.; Pan, X.; Yang, S.Y. Rank Nonnegative Matrix Factorization with Semantic Regularizer for Hyperspectral Unmixing. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2018, 4, 1022–1029. [Google Scholar] [CrossRef]
- Zhang, Z.; Liao, S.; Zhang, H.; Wang, S.; Wang, Y. Bilateral Filter Regularized L2 Sparse Nonnegative Matrix Factorization for Hyperspectral Unmixing. Remote Sens. 2018, 10, 816. [Google Scholar] [CrossRef]
- Shao, Y.; Lan, J.; Zhang, Y.; Zou, J. Spectral Unmixing of Hyperspectral Remote Sensing Imagery via Preserving the Intrinsic Structure Invariant. Sensors 2018, 18, 3528. [Google Scholar] [CrossRef] [PubMed]
- Miao, L.; Qi, H. Endmember extraction from highly mixed data using minimum volume constrained nonnegative matrix factorization. IEEE Trans. Geosci. Remote Sens. 2007, 45, 765–777. [Google Scholar] [CrossRef]
- Cichocki, A.; Zdunek, R. Multilayer nonnegative matrix factorisation. Electron. Lett. 2006, 42, 947–948. [Google Scholar] [CrossRef]
- Rajabi, R.; Ghassemian, H. Spectral unmixing of hyperspectral imagery using multilayer NMF. IEEE Trans. Geosci. Remote Sens. Lett. 2015, 12, 38–42. [Google Scholar] [CrossRef]
- Halimi, A.; Honeine, P.; Bioucas-Dias, J.M. Hyperspectral unmixing in presence of endmember variability, nonlinearity, or mismodeling effects. IEEE Trans. Image Process. 2016, 25, 4565–4579. [Google Scholar] [CrossRef] [PubMed]
- Ghaffari, O.; Zoej, M.J.V.; Mokhtarzade, M. Reducing the effect of the endmembers’ spectral variability by selecting the optimal spectral bands. Remote Sens. 2017, 9, 884. [Google Scholar] [CrossRef]
- Somers, B.; Asner, G.P.; Tits, L.; Coppin, P. Endmember variability in spectral mixture analysis: A review. Remote Sens. Environ. 2011, 115, 1603–1616. [Google Scholar] [CrossRef]
- Costanzo, D.J. Hyperspectral imaging spectral variability experiment results. In Proceedings of the IEEE International Geoscience and Remote Sensing Symposium, Honolulu, HI, USA, 24–28 July 2000. [Google Scholar]
- Zou, J.; Lan, J.; Shao, Y. A Hierarchical Sparsity Unmixing Method to Address Endmember Variability in Hyperspectral Image. Remote Sens. 2018, 10, 738. [Google Scholar] [CrossRef]
- Zhang, H.; Gong, M.; Zhao, D.; Yan, P.; Cui, R.; Jia, W. Super-resolution technique of microzooming in electro-optical imaging systems. J. Mod. Optic 2001, 48, 2161–2167. [Google Scholar] [CrossRef]
- Ricardo, A.; Tales, I.; José, C.; Cédric, R. A Fast Multiscale Spatial Regularization for Sparse Hyperspectral Unmixing. IEEE Trans. Geosci. Remote Sens. Lett. 2018. [Google Scholar] [CrossRef]
- Marian, D.; José, M.; Antonio, P. Total variation spatial regularization for sparse hyperspectral unmixing. IEEE Trans. Geosci. Remote Sens. 2012, 50, 4484–4502. [Google Scholar]
- Matheson, D.S.; Dennison, P.E. Evaluating the effects of spatial resolution on hyperspectral fire detection and temperature retrieval. Remote Sens. Environ. 2012, 124, 780–792. [Google Scholar] [CrossRef]
- Karnieli, A. A review of mixture modeling techniques for sub-pixel land cover estimation. Remote Sens. Rev. 1996, 13, 161–186. [Google Scholar]
- Keshava, N.; Mustard, J.F. Spectral unmixing. IEEE Signal. Process. Mag. 2002, 19, 44–57. [Google Scholar] [CrossRef]
- Torres-Madronero, M.C.; Velez-Reyes, M. Integrating Spatial Information in Unsupervised Unmixing of Hyperspectral Imagery Using Multiscale Representation. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2017, 7, 1985–1993. [Google Scholar] [CrossRef]
- Hay, G.; Niemann, K.; Goodenough, D. Spatial thresholds, image-objects, and upscaling: A multiscale evaluation. Remote Sens. Environ. 1997, 62, 1–19. [Google Scholar] [CrossRef]
- Hao, D.; Xiao, Q.; Wen, J.; You, D.; Wu, X.; Lin, X.; Wu, S. Advances in upscaling methods of quantitative remote sensing. J. Remote Sens. 2018, 22, 408–423. [Google Scholar]
- Rahul, R.; Hamm, N.A.; Kant, Y. Analysing the effect of different aggregation approaches on remotely sensed data. Int. J. Remote Sens. 2013, 34, 4900–4916. [Google Scholar]
- Chai, T.; Draxler, R.R. Root mean square error (RMSE) or mean absolute error (MAE)?—Arguments against avoiding RMSE in the literature. Geosci. Model. Dev. 2014, 7, 1247–1250. [Google Scholar] [CrossRef]
- Nascimento, J.M.P.; Dias, J.M.B. Vertex component analysis: A fast algorithm to unmix hyperspectral data. IEEE Trans. Geosci. Remote Sens. 2005, 43, 898–910. [Google Scholar] [CrossRef]
- Heinz, D.; Chang, C.-I. Fully constrained least squares linear mixture analysis for material quantification in hyperspectral imagery. IEEE Trans. Geosci. Remote Sens. 2000, 39, 529–545. [Google Scholar] [CrossRef]
- Miao, X.; Gong, P.; Swope, S.; Pu, R.; Carruthers, R.; Anderson, G.L.; Heaton, J.S.; Tracy, C.R. Estimation of yellow starthistle abundance through CASI-2 hyperspectral imagery using linear spectral mixture models. Remote Sens. Environ. 2006, 101, 329–341. [Google Scholar] [CrossRef]
- Rahman, A.F.; Gamon, J.A.; Sims, D.A.; Schmidts, M. Optimum pixel size for hyperspectral studies of ecosystem function in southern California chaparral and grassland. Remote Sens. Environ. 2003, 84, 192–207. [Google Scholar] [CrossRef]
- Jia, K.; Liang, S.; Gu, X.; Baret, F.; Wei, X.; Wang, X. Fractional vegetation cover estimation algorithm for Chinese GF-1 wide field view data. Remote Sens. Environ. 2016, 177, 184–191. [Google Scholar] [CrossRef]
- Niroumand-Jadidi, M.; Vitti, A. Reconstruction of River Boundaries at Sub-Pixel Resolution: Estimation and Spatial Allocation of Water Fractions. ISPRS Int. J. Geo-Inf. 2017, 6, 383. [Google Scholar] [CrossRef]




| Accuracy (10−4) | MHS-HU | MLNMF | VCA-FCLS | L1/2-NMF | SUnSAL-TV |
|---|---|---|---|---|---|
| MSEa | 1.073 | 1.093 | 2.178 | 1.102 | 3.274 |
| RMSE | 1.667 | 1.679 | 3.152 | 1.738 | 33 |
| MHS-HU | SUnSAL-TV | |
|---|---|---|
| simulated data | 10.62s | 88.93s |
| Value | 2 | 4 | 5 | 6 | 8 |
|---|---|---|---|---|---|
| MSEa (10−4) | 1.0786 | 1.0284 | 1.0244 | 1.0223 | 1.0227 |
| RMSE (10−4) | 1.8750 | 1.8367 | 1.8376 | 1.8431 | 1.8277 |
| rmsSAD | 0.0736 | 0.0683 | 0.0647 | 0.0672 | 0.0676 |
| 0 | 0.01 | 0.05 | 0.1 | 0.2 | 0.4 | 0.5 | 0.8 | 1 | 2 | |
|---|---|---|---|---|---|---|---|---|---|---|
| MSEa (10−4) | 1.1177 | 1.1197 | 1.1972 | 1.0943 | 1.0742 | 1.0892 | 1.1100 | 1.2264 | 1.3239 | 1.8116 |
| RMSE (10−4) | 1.7948 | 1.7544 | 1.7447 | 1.7534 | 1.7201 | 1.9674 | 2.2948 | 2.7408 | 3.4200 | 5.5022 |
| rmsSAD | 0.0389 | 0.0374 | 0.0385 | 0.0384 | 0.0387 | 0.0428 | 0.0457 | 0.0555 | 0.0621 | 0.0898 |
| Accuracy | MHS-HU | MLNMF | VCA-FCLS | L1/2-NMF | SUnSAL-TV |
|---|---|---|---|---|---|
| rmsSAD | 0.1940 | 0.2329 | 0.2856 | 0.2415 | X |
| RMSE | 0.0063 | 0.0064 | 0.0114 | 0.0088 | 0.0071 |
| Window Scale | 1 | 3 | 5 | 7 |
|---|---|---|---|---|
| rmsSAD | 0.2079 | 0.1953 | 0.1915 | 0.1917 |
| RMSE | 0.0089 | 0.0064 | 0.0061 | 0.0071 |
| SAD | MHS-HU | MLNMF | L1/2-NMF | VCA-FCLS |
|---|---|---|---|---|
| Asphalt | 0.1745 | 0.1692 | 0.2027 | 0.1978 |
| Grass | 0.0896 | 0.0890 | 0.2857 | 0.3667 |
| Tree | 0.1682 | 0.1636 | 0.1897 | 0.2155 |
| Roof | 0.1493 | 0.1569 | 0.4611 | 0.4636 |
| shadow | 0.3289 | 0.3489 | 0.4238 | 0.3881 |
| mean | 0.1821 | 0.1855 | 0.3439 | 0.3263 |
| RMSE | MHS-HU | MLNMF | L1/2-NMF | VCA-FCLS |
|---|---|---|---|---|
| mean | 0.0195 | 0.0198 | 0.03753 | 0.02102 |
| Window Scale | 1 | 3 | 5 | 7 |
|---|---|---|---|---|
| rmsSAD | 0.2398 | 0.2455 | 0.1821 | 0.2424 |
| RMSE | 0.0211 | 0.0185 | 0.0189 | 0.0198 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zou, J.; Lan, J. A Multiscale Hierarchical Model for Sparse Hyperspectral Unmixing. Remote Sens. 2019, 11, 500. https://0-doi-org.brum.beds.ac.uk/10.3390/rs11050500
Zou J, Lan J. A Multiscale Hierarchical Model for Sparse Hyperspectral Unmixing. Remote Sensing. 2019; 11(5):500. https://0-doi-org.brum.beds.ac.uk/10.3390/rs11050500
Chicago/Turabian StyleZou, Jinlin, and Jinhui Lan. 2019. "A Multiscale Hierarchical Model for Sparse Hyperspectral Unmixing" Remote Sensing 11, no. 5: 500. https://0-doi-org.brum.beds.ac.uk/10.3390/rs11050500