1. Introduction
Forests play a key role in the global carbon cycle, storing 80% of all terrestrial aboveground carbon [
1] and contribute substantially to the total annual global terrestrial carbon sink (3.0 ± 0.8 GtC/yr) [
2] with recent estimates placing the net global forest sink at 1.1 ± 0.8 GtC/yr [
3]. Sustainable forest management (SFM) practices could be developed to enhance rates of carbon sequestration in forests (e.g., [
4]). To account for the impact of SFM practices on rates of carbon storage over large areas, spatially comprehensive techniques are needed to quantify carbon stocks (tons/ha) and predict carbon stock changes (tons/ha/yr). This would allow forest managers to (i) make more informed management decisions and; (ii) enhance the use of forests to limit the rate of climate warming [
5].
Predictions of aboveground forest carbon accumulation (i.e., tree growth) are often made by applying statistical models of forest growth and yield (G&Y) to plot-level inventory data [
6]. The key challenge is the extrapolation of plot-level data over larger areas. Plot-scale forest inventories often have limited point sampling, high cost, and limited personnel [
5,
7]. These G&Y models may require inputs of tree-level diameter-at-breast height (DBH), species, site quality, and other attributes to project future carbon accumulation [
8]. To apply G&Y models over large areas, remote sensing methods would be required to collect spatially-explicit (i.e., “wall-to-wall”) data that can be used to initialize G&Y models. Spatially comprehensive estimates of biomass and growth would support SFM, especially in complex forests with multiple tree ages (i.e., size classes) and species [
9].
Light detection and ranging (LiDAR; also referred to as airborne laser scanning; ALS) is an active remote sensing system that generates a 3-D point cloud from which statistical metrics defining vertical and horizontal forest structure are derived [
10,
11]. In this discrete return system, a pulse of laser light travels to the object, where the time of travel to and from the object for each individual laser pulse is recorded to determine the distance, referred to as pulsed ranging [
12]. A discrete-return sensor can transmit up to five returns; when a laser pulse hits the bare ground or a dense object, only a single return is recorded [
11,
13]. Conversely, when the pulse travels through a forest canopy, some component energy can intercept various components of the understory, as well as the ground, resulting in multiple returns [
11]. The metrics, which statistically describe the point cloud (e.g., percentiles, density) can be used as independent variables to predict stem diameter distribution (SDD; i.e., the number of stems/ha across a set of diameter size classes), stem density (SD; stems/ha for a given plot), species abundance, and other attributes needed to initialize G&Y models [
9,
13,
14]. Studies that predict SDD from LiDAR have been limited; this can be partially attributed to the low accuracy of predicting multiple response variables, which complicate model calibration and validation [
14]. SDD is thus often weighted by basal area (BA); i.e., the cross-sectional area of a tree at breast height across a hectare, in m
2/ha) due to the intrinsic quadratic relationship between DBH and BA [
15]; BA distribution (BA_dist) (i.e., the value of the summed individual-tree BA across a hectare in a distributed range, e.g., size classes, in m
2/ha), gives greater importance to larger trees and improves predictions, thus is often used in SDD studies [
16,
17]. Previous studies have initialized local G&Y models using LiDAR-predicted variables to evaluate BA_dist as a proxy for growth [
5], or map present aboveground carbon density using predictions of SDD [
18]. Others have used process-based modelling to evaluate its effectiveness in single-species forests/plantations for BA_dist growth [
19] and biomass [
20]. Zhang et al. (2018) [
21] used repeat LiDAR surveys to calibrate biomass change. Thus, the application of LiDAR to quantify stand characteristics for the initialization of G&Y models has the potential for predicting carbon accumulation in uneven-aged mixedwood temperate forests.
Area-based approaches (ABAs) to quantify SDD and BA_dist using LiDAR metrics include both parametric and non-parametric techniques. Parametric techniques assume that the data conforms to a given probability density function (PDF; [
22]) such as the Weibull distribution, which has been used extensively to predict SDD and BA_dist [
23,
24]. Some have alluded to predicting future growth based on moderate success predicting SDD and other stand attributes across a landscape using parametric approaches (e.g., [
18]). However, modelling of SDD and BA_dist in complex forests has proven to be more challenging. As a result, non-parametric techniques such as
k-nearest neighbours (
k-NN) and random forests (RF) are becoming increasingly popular [
5,
8,
25,
26]. Non-parametric techniques offer the advantage of identifying important variables using variable reduction techniques and eliminating those variables with high collinearity, thus promoting them for LiDAR-based applications in complex forests [
14,
27].
To date, the application of non-parametric approaches to create tree lists and project G&Y models for carbon accumulation has been limited, and, to the best of our knowledge, virtually non-existent in complex forests with mixed species and age structures. For the purposes of this study, we defined tree lists as lists of species-assigned stem diameters for the number of trees in a stand, excluding site class or height. A previous study used
k-NN imputation to parameterize the Forest Vegetation Simulator (FVS; [
28]) over a large forested area in Oregon, finding that
k-NN exhibited higher accuracy for larger trees and concluded that LiDAR could be used for spatially explicit BA_dist projections [
5]. Tompalski et al. (2018) [
29] used an ABA approach to find the best matching yield curves compared to those generated by a locally parameterized model. Others report predictions of single-tree attributes including SDD, but do not compare projections of future growth (e.g., [
14,
30]). Many studies that have used the RF non-parametric algorithm found it stronger at predicting forest attributes than other non-parametric approaches, including
k-NN [
14,
30,
31], however, there remains no consensus on a preferred model across conditions [
8]. Application of RF for predicting forest attributes in complex stands (i.e., mixedwood, uneven-aged) is increasing given it alleviates several challenges associated with parametric techniques, including: (i) the need for significant a priori knowledge to properly parameterize models; (ii) collinearity between predictors; and (iii) overfitting when a large number of predictor variables are used [
14]. Whereas parametric models must be highly parameterized to predict complex SDDs and often require forest type stratification to improve model accuracy, RF is not constrained by these requirements [
24,
31,
32,
33]. Predicting species abundance, a requirement in initializing many G&Y models, has also proved to be challenging in mixedwood forests due to increased structural complexity, coupled with the challenges of accurately normalizing LiDAR strength of return (i.e., intensity [
34,
35]), although advances have been made (e.g., [
36,
37]). However, the error associated with an imprecise or inaccurate characterization of species abundance on growth estimates remains unclear, as studies do not typically compare any potential changes in accuracy using inventory-derived species abundance compared to predictions from LiDAR for G&Y models (e.g., [
5,
38,
39]).
In general, predicting tree growth, specifically using ABAs, has a number of challenges due to the need to disaggregate size class-level predictions to tree-level, apply this across a larger area, and then impute or otherwise obtain species abundance. In this study, we tested the initialization of a LiDAR-derived G&Y model using average diameter tree lists combined with a suite of G&Y model specifications and tests of different approaches to quantify species abundance. We used estimates of BA_dist and SDD/SD to initialize an individual tree-based G&Y model (i.e., FVSOntario) to investigate the feasibility of forecasting carbon accumulation (i.e., the difference in carbon stock between years) in an uneven-aged, mixedwood forest (i.e., the Great Lakes - St. Lawrence forest). Our objectives were: (1) initialize a baseline G&Y model (i.e., FVS1) using plot-based diameter-at-breast-height (DBH) measures and species abundance data; (2) initialize a second G&Y model (i.e., FVS2) based on LiDAR-derived size-class DBH estimates and plot-based species abundance data; (3) initialize a third model (i.e., FVS3) using LiDAR-derived size-class DBH estimates and species abundance data inferred from the Forest Resource Inventory (FRI); (4) initialize a fourth model using BA_dist and median size-class range values to evaluate the effect of modelling without SDD (i.e., FVS3-2), and (5) compare predicted carbon accumulation for the different models to inventory-based estimates of carbon accumulation. We compared stocks at model initialization to 2012 inventory validation data (i.e., FVS1), projected stocks for all the models after a four-year simulation to 2016 inventory validation data, and further compared LiDAR model outputs at 2021.
3. Results
3.1. Deriving Tree Lists
Differences between RMSE and RMSEcv were small, indicating the SDD, SD, and BA_dist models were stable (
Table 4). BA_dist was predicted with moderate accuracy, with plot-level predictions and medium and above sawlogs having the highest predictive accuracy in the model validation. rRMSE was lower for BA_dist than SDD/SD, indicating that BA_dist was better predicted relative to the mean value. However, both predictions had higher sRMSD (i.e., higher standard deviations of errors). In general, BEI was high, i.e., 0.84, 0.67, 0.34, 0.75, 0.76, and 0.64, for PIN, THW, INT, CU, CL, and MX, respectively (0.73 averaged over the four size classes). Observed and predicted values of BA_dist and SD/SDD by forest type are presented in
Figure 5 and
Figure 6 respectively.
We omitted the FVS3-2 model from the further discussion due to poor results. To investigate LiDAR-projected carbon accumulation values compared to FVS1 projections, additional comparisons were made to FVS1 for both carbon stocks and accumulation at the plot and size-class level to investigate any statistical equivalence; none of the models showed a statistical equivalence of proportionality (p > 0.05) for either mortality rate, although some had equivalent means. Additional comparisons were made for carbon accumulation. Interestingly, while we did find that the means yielded equivalent results at both mortality rates, proportionality remained underpredicted by the LiDAR-based models.
3.2. G&Y Modelling for Carbon Stocks at Model Initialization and Four-Year Projection
When compared to initial (2012) validation carbon stocks (i.e., FVS1 initialization), we found low correlation across all plot-level models and generally high deviations from the mean and spread of variation (
Table 5). The plot-level model with the highest correlation was FVS3 aggregate (R = 0.16), slightly higher than the FVS2 aggregate equivalent (R = 0.12); this stayed relatively consistent when projected to 2016. LiDAR-initialized models generally had a low bias, while aggregate and average models had similar bias across inventory versus FRI species models.
The equivalent FVS average models were not correlated for 2012 or 2016. None of the models were proportionally equivalent to inventory carbon stocks but were statistically unbiased. FVS2 and FVS3 aggregate models had high heteroscedasticity in observed versus predicted values (
Figure 7), becoming increasingly variable at higher carbon stocks; this did not stabilize in 2016 (
Figure 8). FVS1-projected was statistically equivalent to inventory measures of carbon stocks for 2016 for bias and proportionality at both constant and species-specific mortality rates (R = 0.99, 0.98, respectively). There was no statistical difference between projected values of carbon stock in 2016 for either mortality scenarios for LiDAR-initialized models, referring to the range of CIs within proportionality.
At a size-class level, projections varied widely across both years (
Table 6). None of the LiDAR-initialized size-class models were proportionally equivalent to the inventory carbon stocks for either year and were generally highly skewed from the mean and had a high spread of variance; models remained stable in 2016. Mortality rates had negligible effects on the overall model accuracy and bias.
FVS1 was equivalent in both bias and proportionality to 2016 inventory carbon stocks, except for the polewood size class at constant mortality where proportionality was closer to the 0.0−0.3 range (i.e., consistently underpredicting) where a value of 0.75 is the minimum CI range considered equivalent. FVS1 saplings at both mortality rates were proportionally equivalent but not equivalent in bias.
On a forest-type level, there were some variances in correlation in 2012, especially for the CU group (n = 8; R = 0.70 for FVS2 Aggregate; R = 0.76 for FVS3 aggregate). Generally, groups were consistent with overall results, with a high spread of variation that ranged from rRMSE = 0.39 for FVS3 aggregate in the INT group (n = 6), to 2.42 in the FVS2 aggregate CL group (n = 9). For 2016, these correlations remained relatively consistent (CU FV2 aggregate R = 0.73 at constant mortality; FVS3 aggregate R = 0.79 at both mortality rates). For size classes, forest types still had high deviations from observed values at both years. Carbon stocks for CU medium and above sawlogs had a moderate correlation with inventory data for both inventory and photo-interpreted (FRI) species models in 2012 (R = 0.42, 0.43, respectively), however, these values were extremely far from the observed mean and had a high spread of variance (rRMSEmax = 12.91; sRMSDmax = 6.77). THW small sawlogs had similar correlations (i.e., R = 0.37 and 0.34, for FVS2 and FVS3, respectively), with lower deviations from the mean and lower spread of variance (rRMSEmax = 0.83; sRMSD%max = 1.01). INT saplings had a higher correlation for FVS3 at R = 0.57, and had a similar trend in mean and spread of variation, at rRMSE = 0.94 and sRMSD = 2.88.
3.3. G&Y Modelling of Carbon Accumulation (2012 to 2016)
Carbon accumulation predictions were highly biased and had high rRMSEs and sRMSDs (
Table 7). In contrast to carbon stock projections, carbon accumulation was not statistically equivalent for the baseline FVS1-projected model for either bias or proportionality to measured inventory-based accumulation at either mortality rate, where a constant mortality rate generally overpredicted, and a species-specific mortality rate underpredicted mean accumulation (
Figure 9).
Correlation between LiDAR-initialized model values and inventory rates were comparable to 2012/2016 carbon stock projections (e.g., 2012 FVS2 aggregate R = 0.16; 2016 R = 0.11; accumulation constant mortality R = 0.13, species-specific mortality = 0.17), with comparable spreads of variation (i.e., sRMSD), however, predictions were considerably higher than the observed means (i.e., higher rRMSE). Compared to predictions of carbon stock, accumulation predictions were either largely systematically over or under-predicted. Accumulation appeared consistent across both FVS1 and LiDAR-initialized models, with the exception of a few outliers for FVS1 (
Figure 10).
At the size-class level, none of the models were proportionally equivalent to inventory-based accumulation, although some models were statistically equivalent to the mean (i.e., the models were equivalent in bias). RMSE was still high for all groups, although some models had high correlations and similar distributions of values visually.
On a forest type level, some incremental improvements in correlation and error were observed, while some models were several orders of magnitude under- or over-predicted versus the mean observation. Specifically, the accumulation from INTconstant, CLconstant, and CLspecies-specific forest types all had rRMSEs greater than ±5.00. The strongest predictions were for the PIN type, with the FVS1 mean approaching an rRMSE of 0.78 for constant mortality and 1.01 for species-specific mortality, comparable to 0.78 for the FVS3 aggregate constant mortality model, and 0.88 for the species-specific model; bias was generally positive for constant mortality (FVS1 bias = 15.5%) and negative for species-specific mortality (FVS1 bias = −41.13%).
3.4. Influence of Species Designation on Carbon Stock Predictions
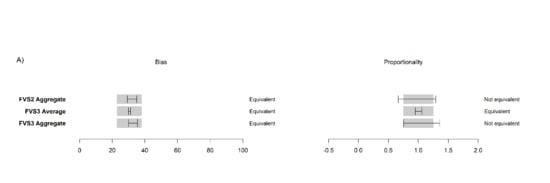
At the plot-level, FVS2 was statistically equivalent to FVS3 for 2012, 2016, and a nine-year projection at both mortality rates, shown here for FVS2 average at constant mortality (
Figure 11,
S4).
At the size-class level, predictions were more varied. Medium and above sawlogs were not equivalent in proportionality (p > 0.05) for 2012, 2016, or 2021, however, they were equivalent in bias for both years. In contrast, small sawlogs and polewoods were found to be equivalent in bias and proportionality in all three years at both mortality rates for FVS2 to FVS3 equivalent models. Testing constant mortality, carbon stock projections grouped by forest types held equivalency for 2016 except for the INT and MX types. We did not test by size class due to the insufficient number of plots needed to perform a meaningful statistical equivalence test.
3.5. Influence of Species Designation on Carbon Accumulation Predictions
Carbon accumulation was generally not equivalent when compared between LiDAR-initialized models at the plot level. FVS2 aggregate models were equivalent to FVS3 aggregate models at the constant mortality level for 2016 and 2021, but not for species-specific mortality. Similar trends were observed at the size class level. Medium and above size class accumulation rates were equivalent for 2016 and 2021 for constant mortality, but were not equivalent for species-specific mortality, while saplings were not equivalent at either rate at either year. Small sawlogs and polewoods were equivalent at both years in both bias and proportionality for constant mortality, but not for species-specific mortality. Accumulation by forest type (i.e., PIN, THW, INT, CU/CL, MX) did not retain proportional equivalency for constant mortality for 2016 and thus was not examined for 2021.
4. Discussion
To the best of the authors’ knowledge, no study has projected carbon accumulation in mixed temperate stands using local G&Y models from LiDAR-initialized tree lists and photo-interpreted species abundance data. We determined the utility of LiDAR-initialized G&Y models to improve estimates of carbon accumulation and carbon stocks in an understudied mixedwood temperate forest compared to projections using inventory data. We also sought to understand the importance of precise species abundance data on predictions of carbon accumulation and carbon stock using a locally-tailored model.
Equivalence testing indicated that neither FVS2- nor FVS3-based carbon accumulation or carbon stocks at any size class and at the plot level were equivalent to inventory-initialized estimates at a four and nine-year projection, emphasizing the difficulty in parameterizing models for complex temperate forests with moderate-to-poor SDD/SD and BA_dist predictions. Inventory-based FVS1 predictions were equivalent for carbon stocks, but failed to maintain equivalency for 2012−2016 accumulation compared with validation data, signifying that accumulation is more sensitive to precise parameterization than carbon stocks. A key finding was that FVS2 and FVS3 (i.e., inventory-based measures of species abundance and photo-interpreted species, respectively) were equivalent for most size classes and at the plot level (
p < 0.05). They were also generally equivalent when examined at a forest type level for carbon stocks, but not for accumulation. This finding suggests that predictions of species abundance from LiDAR (e.g., [
5,
81]) may possibly be replaced in studies where photo-interpreted estimates are available for the purpose of improving carbon stock estimates and broad accumulation estimates. Testing mortality rates showed that the constant mortality rate was more effective due to the high variability that species-specific rates caused, especially for photo-interpreted parameterization. Furthermore, these data are spatially-explicit and can be used in conjunction with LiDAR data to initialize tree-lists and map carbon accumulation at the forest level.
4.1. Assessment of BA_dist, SDD, and SD Attribute Predictions
Model predictions of BA_dist were better than SDD and SD predictions. Results were favourable in comparison to other studies that predicted BA_dist from LiDAR in similar forest types; i.e., Spriggs et al. (2017) [
18] reported an rRMSE of 22% and 33% in sugar maple and mixedwood plots, respectively (plot size = 2500 m
2), Vauhkonen & Mehtätalo (2014) [
82] reported an rRMSE of 13–68% (Scots Pine plantations; plot size = 400 m
2), Lamb et al. (2017) [
83] reported an rRMSE of 36.6% in spruce plantations; compared to 29% in this study.
Falkowski et al. (2010) [
5] used a
k-NN/RF imputation approach to predict SDD/SD and BA_dist in a conifer-dominated forest, achieving a correlation of 0.88, considerably higher than our findings (R = 0.62), while their best-performing SD model performing similarly to ours (R = 0.29; R = 0.28, respectively). Our small trees had lower correlations compared to their best sapling model (R = 0.49, compared to R = 0.32 in this study) although our definition of sapling was more constrained (i.e., 8–17 cm DBH, versus 8–18 cm). Although not evaluated in this study, they also reported that none of the measured forest inventory variables, including total BA_dist and SD, were proportionally equivalent to validation data at the ±20% equivalence level (
p < 0.05). Spriggs et al. (2017) [
18] discussed the difficulty in predicting smaller stems, particularly in closed canopies where understory returns are limited. Hudak et al. (2008) [
25] reported a high correlation (R = 0.80) for BA_dist in a similar mixed conifer forest, comparable to the results reported here (R = 0.79). Using an ITC approach, Lindberg et al. (2010) [
84] achieved rRMSEs of 37% for SD in a mixedwood forest, close to the 43% reported here, while Vauhkonen and Mehtätalo (2014) [
82] reported an rRMSE of 27.6% for SD.
RF is a common non-parametric algorithm used in forestry attribute prediction [
75]. The challenge encountered in this study was the difficulty in accurately predicting SDD and BA_dist values when there was an absence of trees for a given size class. This was most pronounced for medium and above trees across all forest types resulting in an over-prediction of carbon stock values for a considerable proportion of plots. Here, we saw a consistent bias in our observed and predicted values, where a considerable number of plots had no medium or above diameter trees, but the RF model for BA_dist and SDD predicted the presence of trees. This could be due to the inherent characteristic of RF to average predicted values, where a prediction of zero is unlikely in RF regression trees. Shang et al. (2016) [
85] suggested that in attribute prediction, larger size classes could be pooled together to improve model performance of SDD predictions. Following this directive, sRMSD for medium and above size classes was 59%, considerably lower than the sRMSD reported for their 38–50, 50–62, and 62 and above size classes (sRMSD = 78, 80, and 89%, respectively). However, we found that following this protocol ultimately did not translate to statistically equivalent measures of carbon stock or accumulation to inventory data.
The SDD model selected intensity metrics for smaller size classes, and height-based metrics for larger size classes. In similar studies where intensity metrics have been used, they have also been selected as important for smaller size classes [
14]. Although intensity helped in predicting these small size classes (as measured by %IncMSE), it ultimately did not lead to proportional equivalency for carbon stocks or accumulation. Conversely for BA_dist, only height variables were selected and still resulted in a considerable decrease in G&Y model accuracy (i.e., FVS3-2).
4.2. Assessment of G&Y Models
Again, to our knowledge no studies have forecasted carbon accumulation using a predicted average-DBH tree list from LiDAR. Some studies have compared remeasurement data from repeat LiDAR surveys to derive carbon accumulation; Hudak et al. (2012) [
76] used repeat LiDAR surveys to quantify carbon accumulation over a six-year period by subtracting LiDAR-predicted biomass, and Økseter et al. (2015) [
86] used similar techniques for an 11-year period. In general, our LiDAR-initialized models projecting carbon stock were not equivalent to the inventory stocks in proportionality, regardless of the type of plot-level tree list initialization (i.e., using plot-level SD and BA_dist, or using an aggregated list from size-class level predictions) or whether looking at the size-class level. However, aggregate models had a consistently higher spread of values and a consistent underprediction of larger carbon stock plots than average models. Accumulation was more sensitive; our LiDAR-initialized models consistently over- or under-reporting accumulation rates, indicating mortality was not ideally parameterized and should be more carefully considered. Perhaps the addition of an ‘average’ rate could have improved the mean accumulation results. Since results were similar regardless of species abundance data used and regardless of mortality, it implies that further refinement is needed in creating accurate DBH for G&Y model initialization and that perhaps using a distribution of DBH values may improve modeling results. Indeed, perhaps more information is needed that goes beyond the ability of LiDAR to provide for carbon stocks or accumulation, and that integration of more ancillary data is necessary, such as aerial photography, Sentinel-2 A or other remotely-sensed data to provide stronger initial estimates of BA_dist and SD/SDD prior to G&Y modeling [
87,
88].
Our findings indicate an agreement of carbon stock values for FVS1 at the plot and size-class level with validation data (except for saplings). However, none of the FVS1-projected accumulation models were equivalent to inventory measures. Similarly, Hudak et al. (2012) [
76] reported more variability in accumulation when extracted from biomass predictions. Accumulation appeared to be in agreement when looking at observed versus predicted values between FVS1 and LiDAR-based models, with similar rRMSEs (e.g., rRMSE
PIN = 77.8% for FVS1; 86.50% for FVS2 aggregate at constant mortality), suggesting that some mechanistic growth patterns were consistent (
Figure 6). Because our subsequent equivalence testing indicated the models were still dissimilar proportionally, we stratified the plot-level accumulation models by forest type to identify any consistencies with FVS1 projections, however, this further reduced the accuracy of accumulation results from LiDAR models and means became statistically dissimilar, in addition to the already statistically dissimilar proportionalities. Looking at the highest deviations in plot-level accumulation across model types (
Figure 6), we examined five plots that were clear visual outliers, two of which were consistent across model types. For constant mortality, plot number 026, an intolerant hardwood type consisting mainly of
B. papyrifera,
P. grandidentata,
A. rubrum, and
A. saccharum had a predicted accumulation rate of 0.9 Mg/ha/yr, compared to an actual rate of 2.24 Mg/ha/yr. For FVS1, these extreme deviations typically occurred at species-specific mortality rates and within CU and INT groups (e.g., plot number 075 predicted 0.34 Mg/ha/yr versus −2.28 Mg/ha/yr CU; plot number 026 −0.36 Mg/ha/yr versus −2.24 Mg/ha/yr INT). Studies have previously identified conifers as being difficult to capture with LiDAR due to the reflective properties of crowns, however, these inaccuracies were also present in the inventory model [
18]. Given that these outliers were generally more pronounced with species-specific mortality rates, it is likely that our models did not parameterize ideal rates for specific hardwoods and that future consideration of mortality rate should be more carefully reviewed.
We did not expect to see statistical equivalencies for carbon stock and accumulation forecasts for FVS2 and FVS3 (meanFVS2STOCK2016_CONST = 33.48 tons/ha, meanFVS3STOCK2016 = 33.74 tons/ha, meanFVS2ACCUMULATION_CONST = 3.22 tons/ha/yr, meanFVS3ACCUMULATION_CONST = 3.22 tons/ha/yr).The majority of photo-interpreted species data was from 2009, with a smaller proportion from 2014. This two-to-three-year age gap between LiDAR and species abundance data collection suggests that the species composition played less of a role in initializing the G&Y models compared to DBH, even for a G&Y model that includes species-specific allometric equations. This emphasizes the need for a renewed call on accurate inputs of DBH as the primary driving factor of a more accurate carbon accumulation forecast. We also expected less of an agreement as the model was projected into 2021, however, the LiDAR-based models still retained statistical equivalency for specific combinations; i.e., accumulation models retained equivalency for the aggregate model with constant mortality over the nine-year scenario, suggesting that better predictions of larger size classes influenced the consistency of carbon accumulation predictions. We did find some discrepancies between LiDAR-based models for certain size classes. Because saplings grow at a faster rate, we did not expect photo-interpreted species to be proportionally equivalent as growth occurs at a faster rate for young trees, thus precise parameterization of species would have more influence here. There was a closer agreement between FVS2 and FVS3 at species-specific mortality for medium and above carbon stocks; however, this size class only retained accumulation equivalency at a constant mortality rate. Because a species-specific mortality rate coupled with an imprecise species characterization could further skew values, the use of the constant mortality rate is preferred. Ultimately, future work could also compare the results of forest attribute modeling with LiDAR-derived species abundance and photo-interpreted estimates for potential model improvement.
As species abundance is data and resource-intensive to calibrate due to the high range of forest conditions needed to be captured [
89], it may be prohibitive to initialize models based on remotely-sensed predictions. Previous studies at PRF have attained varying levels of success in species abundance modelling (e.g., classification accuracies of 64%
P. strobus, 53.7%
Q. rubra, 79%
P. resinosa, 49%
B. papyrifera, 79.9%
A. saccharum, 33.5%
A. rubrum, 51%
P. glauca, Gougeon and Leckie, 2011 [
90], using an ITC approach; R = 0.69, 0.74, 0.85, 0.53, 0.73 for
P. strobus,
P. resinosa,
P. glauca,
P. betula/
populus,
Acer spp., van Ewijk et al., 2014 [
81], using boosted regression trees [BRTs]). Falkowski et al. (2010) [
5] found that predicting species abundance did not produce statistically equivalent results to inventory measurements (
p < 0.05), however, projections of BA_dist initialized using FVS still followed similar trends to inventory-initialized projections. Thus, using photo-interpreted species abundance may be preferred in complex forests where individual-tree species information is required for G&Y model initialization and for which these data are available. Falkowski et al. (2010) [
5] also demonstrated that it may be possible to achieve statistically equivalent results of growth when using non-inventory-equivalent predicted species abundance with BA_dist projections, rather than estimating tree lists and explicitly projecting carbon stock values. This also suggests that the allometric equations we used in converting diameter to carbon stock may introduce large amounts of error, thereby requiring further refinement.
The removal of the less accurate SDD in predictions and using only BA_dist (i.e., FVS3-2) resulted in statistically different models on average that were less effective than the combined SDD/BA_dist plots, implying that regardless of the low SDD accuracy, it remained a key component in obtaining more accurate tree lists. However, other attributes, such as quadratic mean diameter (QMD) could also be used instead of SDD to further improve predictions, given that QMD has shown improved predictions compared to SDD (e.g., [
9]). Another alternative could be using a predicted distribution of values, capturing a higher range of diameters than the mean (i.e., PDFs; Olofsson et al., 2008 [
91]). Unpublished work by van Ewijk (2015) [
92] used height-diameter relationships to estimate diameters from LiDAR-derived height distributions, albeit discovering that this characterization was not sufficient for generating accurate tree lists for FVS
Ontario initialization.
Given that we had plots with negative accumulation for some years in the validation data, we additionally tested whether the removal of negative mortality would improve model estimates for both inventory and LiDAR-based models using a constant mortality rate. We found that this did not make a large impact on predicted values and that both the inventory and LiDAR-based models were still not equivalent to the validation data for 2016. This suggested that there were additional underlying mechanisms not captured by the G&Y model.
4.3. Limitations and Future Work
Various considerations in this study may have contributed to errors that propagated throughout the modelling effort. One limitation relates to plot size, which was relatively small (i.e., 625 m
2); for instance, Shang et al. (2016) [
85] achieved a BEI of 0.49 for 400 m
2 plots versus a BEI of 0.39 for 1000 m
2 plots. Given our BEI of 0.73, increasing plot size may have decreased our attribute prediction error so long as plot number is not reduced [
93]. It is well-established that a larger plot would reduce the ‘edge effect’, or the difference between field-based plot and LIDAR-based characterization of trees in the same radius (i.e., nature of the canopy captured in the point cloud). For instance, a tree right outside the plot boundary but leaning in towards the plot would be excluded from any mention in inventory data; however, the canopy itself would still be captured in the equivalent LiDAR return, thereby influencing LiDAR metrics and the prediction of variables [
11,
84]. Because forests are complex and the full range of forest dynamics is not yet understood, a modelling approach incorporating a stochastic element could have generated more amenable results [
94]. In addition, a larger number of plots would have enabled us to examine more relationships between model parameterizations and predictive success, for instance further comparing carbon stock and accumulation by forest types at each size-class level.
As with any modelling exercise, a variety of conditions and sensitivities may or may not be captured to characterize the full range of effects. Our data were stratified by common forest types of Ontario based on species abundance ranges in our data; however, stratification by NDVI, forest structural groups, aspect, slope, elevation, etc., may have enhanced model performance [
75].
While we tested for mortality across all modelling approaches, our value for species-specific mortality rate was chosen somewhat arbitrarily based on the differences between 2012 and 2016 live trees. Because FVS
Ontario had a limit of maximum stand stocking density [
95], using the model-supplied internal mortality logic would often result in a massive die-off of trees due to our sometimes-high predictions of stems, thus we were constrained to a user-defined rate. As our defined mortalities often resulted in either under- or over-predictions of LiDAR-based carbon stock and accumulation, it is recognized as an important parameter. Future work should strive to improve the methodology for selecting the most appropriate mortality rate for G&Y models especially when used with LiDAR-initialized tree lists.
At the same time, other studies examine plantations consisting of commercial tree species, rather than a range of dynamic conditions such as in PRF. Boisvenue et al. (2019) [
96] highlight this problem, noting that often, commercial tree species are heavily favoured in sampling, and the use of allometric equations from a limited number of samples often leads to increased error, compounded with the fact that the allometric equation itself may affect estimation of carbon stocks/accumulation. PRF contains a multitude of stem sizes and classes, the smallest of which (<8 cm) were not included in this study, partially due to the assumption that the relative total BA_dist of these trees makes them negligible in carbon accumulation, if not for the fact that their height and relative width make them difficult for LiDAR to capture. We set a height threshold of 1.37 m for this study, below which returns were assumed to be ground returns and ignored in metric calculation. However, smaller (i.e., younger) trees grow faster than large (i.e., older) trees and thus accumulate carbon at a faster rate [
88]. The exclusion of these trees in our inventory and in subsequent modelling could be a missing component that contributes to predicted carbon accumulation (i.e., under-estimating carbon accumulation). However, capturing the true proportion of BA_dist and SD/SDD contained within this size class remains challenging given the lack of resources to collect during field campaigns. If a sampled plot contains only one large tree and an abundance of <8 cm DBH trees, it is plausible that these trees accumulate a total amount of carbon large enough for carbon accounting and reporting. There may be a relationship between the proportion of BA_dist in <8 cm trees and canopy metrics that can be explored in a future study to capture the fullest extent possible of the aboveground live carbon domain. Similar work has been reported by Maltamo et al. (2004) [
39], using tree height distribution as a proxy for estimation of smaller stems, finding poor accuracy for these suppressed trees.
Although beyond the extent of this modelling exercise, Penner et al. (2015) [
31] and Breidenbach et al. (2008) [
97] suggested that there are possible improvements in SDD predictions at larger stand or forest levels; extrapolating the FVS
Ontario model over a larger area may improve predictions of carbon accumulation in a future study, albeit noting the model would need to be modified to allow for spatially-explicit modelling. Previous studies with access to spatially parameterized G&Y models have done this for SD and BA_dist (e.g., [
5,
89,
98]).
Many studies have successfully implemented LiDAR-derived metrics for forest attribute modeling [
25,
75,
87,
99,
100]. Recent studies have also used LiDAR for mapping aboveground carbon density [
23] or using repeat LiDAR surveys to map biomass and establish accumulation rates based on these differences [
76,
101]. However, without the access of repeat LiDAR for future projections, accumulation is usually implied from rates of BA_dist growth, which limits our ability to fully report on carbon sequestration and gain a more holistic sense of Canada’s potential for sequestration in understudied complex temperate forests. Looking ahead to the future of LiDAR and carbon management, the new successfully launched Global Ecosystem Dynamics Investigation (GEDI) is gaining considerable attention [
102]. Given this study’s finding that photo-interpreted estimates from various years can provide equivalent estimates to inventory-level data, future studies should also look to investigate the application of space-based LiDAR and coincident species abundance data for areas with less access to ground data to improve estimates of carbon accumulation.
5. Conclusions
Lidar has been shown to be a valuable and reliable tool in forestry for spatially-explicit modeling of forest stand attributes (e.g., [
9,
10,
16,
18,
25]). In the context of G&Y modelling, LiDAR-derived tree lists for initializing G&Y models of direct carbon stock and accumulation have not been tested, which could lead to new insights for climate change mitigation in forest management practices. Complex temperate forests remain understudied in G&Y research; improving G&Y techniques in this area could benefit climate change mitigation and sustainable forest management. This study examined methods for improving estimates of carbon accumulation and carbon stock using a LiDAR-derived G&Y model in an uneven-aged, mixedwood forest of Ontario, Canada. Our predictions of forest variables using the non-parametric RF algorithm were found to be comparable to other studies, however, small-diameter trees, particularly saplings and polewoods, were less well predicted by LiDAR. We demonstrated that predicting carbon stocks across a range of size classes and forest types were more accurate than predicting carbon accumulation, but neither model output achieved statistical equivalence across a range of forest types compared to inventory data. Thus, the information demands for accurate G&Y modeling of carbon accumulation in complex stands remain beyond the reach of current LiDAR and nonparametric modeling approaches. However, our findings suggest this gap is also due to high parameterization costs of the G&Y model, such as mortality, and that these remain areas of focus.
Statistical equivalence was achieved between LiDAR-based models, suggesting complex forests could benefit from photo-interpreted estimates to achieve the same results as inventory-derived species abundance, possibly bypassing the need for predicting species information in carbon accumulation studies. However, this area requires further study, including why these consistencies do not hold across forest types. Models tended to perform better using constant mortality rates when initialized with photo-interpreted species, which could be explained by the need to balance imprecise species with a more generalized mortality rate. The potential of using photo-interpreted species abundance estimates for areas where such data exists as a potential replacement or comparison for LiDAR-derived species abundance is a novel contribution in this study.
This study explored the various considerations for establishing a G&Y model in the understudied uneven-aged mixed forests of Ontario. Models are inherently deficient due to the complexity of the systems studied; thus assumptions can be flawed. Additionally, it is important to note that field data may not provide any more “true” estimates than LiDAR models, and come with their own inherent measurement and sampling errors. Future studies should continue to explore what parameters are crucial for improved estimates of complex forest carbon accumulation.