Classification of Arrhythmia by Using Deep Learning with 2-D ECG Spectral Image Representation
Abstract
:1. Introduction
1.1. Related Works
1.2. Our Contributions
- Spectrograms (2-D images) are employed, which are generated from the 1-D ECG signal using STFT. In addition, data augmentation was used for the 2-D image representation of ECG signals.
- A state-of-the-art performance was achieved in ECG arrhythmia classification by using the proposed CNN-based method with 2-D spectrograms as input.
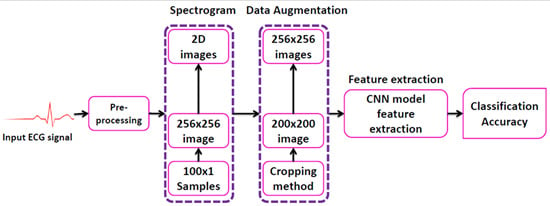
2. Proposed Scheme
2.1. Pre-Processing
2.2. Generation of 2-D Images
2.3. Data Augmentation
2.4. Deep Neural Network
3. Experiments
3.1. Dataset
3.2. Deep Neural Network Parameters
3.3. Experimental Setup
3.4. Cost Function
3.5. Evaluation Parameters
4. Classification Results and Discussion
4.1. Results
4.2. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Mc Namara, K.; Alzubaidi, H.; Jackson, J.K. Cardiovascular disease as a leading cause of death: How are pharmacists getting involved? Integr. Pharm. Res. Pract. 2019, 8, 1. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lackland, D.T.; Weber, S.M.A. Global burden of cardiovascular disease and stroke: Hypertension at the core. Can. J. Cardiol. 2015, 31, 569–571. [Google Scholar] [CrossRef] [PubMed]
- Mustaqeem, A.; Anwar, S.M.; Majid, M. A modular cluster based collaborative recommender system for cardiac patients. Artif. Intell. Med. 2020, 102, 101761. [Google Scholar] [CrossRef] [PubMed]
- Irmakci, I.; Anwar, S.M.; Torigian, D.A.; Bagci, U. Deep Learning for Musculoskeletal Image Analysis. arXiv 2020, arXiv:2003.00541. [Google Scholar]
- Anwar, S.M.; Majid, M.; Qayyum, A.; Awais, M.; Alnowami, M.; Khan, M.K. Medical image analysis using convolutional neural networks: A review. J. Med. Syst. 2018, 42, 226. [Google Scholar] [CrossRef] [Green Version]
- Gu, J.; Wang, Z.; Kuen, J.; Ma, L.; Shahroudy, A.; Shuai, B.; Liu, T.; Wang, X.; Wang, G.; Cai, J.; et al. Recent advances in convolutional neural networks. Pattern Recognit. 2018, 77, 354–377. [Google Scholar] [CrossRef] [Green Version]
- Wu, Y.; Yang, F.; Liu, Y.; Zha, X.; Yuan, S. A comparison of 1-D and 2-D deep convolutional neural networks in ECG classification. arXiv 2018, arXiv:1810.07088. [Google Scholar]
- Zhao, J.; Mao, X.; Chen, L. Speech emotion recognition using deep 1D & 2-D CNN LSTM networks. Biomed. Signal Process. Control 2019, 47, 312–323. [Google Scholar]
- Ortega, S.; Fabelo, H.; Iakovidis, D.K.; Koulaouzidis, A.; Callico, G.M. Use of hyperspectral/multispectral imaging in gastroenterology. Shedding some–different–light into the dark. J. Clin. Med. 2019, 8, 36. [Google Scholar] [CrossRef] [Green Version]
- Feng, Y.-Z.; Sun, D.-W. Application of Hyperspectral Imaging in Food Safety Inspection and Control: A Review. Crit. Rev. Food Sci. Nutr. 2012, 52, 1039–1058. [Google Scholar] [CrossRef]
- Lorente, D.; Aleixos, N.; Gómez-Sanchis, J.; Cubero, S.; García-Navarrete, O.L.; Blasco, J. Recent Advances and Applications of Hyperspectral Imaging for Fruit and Vegetable Quality Assessment. Food Bioprocess Technol. 2011, 5, 1121–1142. [Google Scholar] [CrossRef]
- Tatzer, P.; Wolf, M.; Panner, T. Industrial application for inline material sorting using hyperspectral imaging in the NIR range. Real-Time Imaging 2005, 11, 99–107. [Google Scholar] [CrossRef]
- Kubik, M. Chapter 5 Hyperspectral Imaging: A New Technique for the Non-Invasive Study of Artworks. Phys. Tech. Study Art Archaeol. Cult. Herit. 2007, 2, 199–259. [Google Scholar]
- Hassan, H.; Bashir, A.K.; Abbasi, R.; Ahmad, W.; Luo, B. Single image defocus estimation by modified gaussian function. Trans. Emerg. Telecommun. Technol. 2019, 30, 3611. [Google Scholar] [CrossRef] [Green Version]
- Ahmad, M.; Bashir, A.K.; Khan, A.M. Metric similarity regularizer to enhance pixel similarity performance for hyperspectral unmixing. Optik 2017, 140, 86–95. [Google Scholar] [CrossRef]
- Salem, M.; Taheri, S.; Yuan, J.S. ECG arrhythmia classification using transfer learning from 2-dimensional deep CNN features. In Proceedings of the 2018 IEEE Biomedical Circuits and Systems Conference (BioCAS), Cleveland, OH, USA, 17–19 October 2018; IEEE: Piscataway, NJ, USA, 2018; pp. 1–4. [Google Scholar]
- Mustaqeem, A.; Anwar, S.M.; Khan, A.R.; Majid, M. A statistical analysis based recommender model for heart disease patients. Int. J. Med. Inform. 2017, 108, 134–145. [Google Scholar] [CrossRef]
- Anwar, S.M.; Gul, M.; Majid, M.; Alnowami, M. Arrhythmia Classification of ECG Signals Using Hybrid Features. Comput. Math. Methods Med. 2018. [Google Scholar] [CrossRef] [Green Version]
- Mustaqeem, A.; Anwar, S.M.; Majid, M. Multiclass classification of cardiac arrhythmia using improved feature selection and SVM invariants. Comput. Math. Methods Med. 2018. [Google Scholar] [CrossRef] [Green Version]
- Mustaqeem, A.; Anwar, S.M.; Majid, M.; Khan, A.R. Wrapper method for feature selection to classify cardiac arrhythmia. In Proceedings of the 2017 39th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Jeju Island, Korea, 11–15 July 2017; IEEE: Piscataway, NJ, USA, 2017; pp. 3656–3659. [Google Scholar]
- Minami, K.I.; Nakajima, H.; Toyoshima, T. Real-time discrimination of ventricular tachyarrhythmia with Fourier-transform neural network. IEEE Trans. Biomed. Eng. 1999, 46, 179–185. [Google Scholar] [CrossRef]
- Coast, D.A.; Stern, R.M.M.; Cano, G.G.; Briller, S.A. An approach to cardiac arrhythmia analysis using hidden markov models. IEEE Trans. Biomed. Eng. 1990, 37, 826–836. [Google Scholar] [CrossRef]
- Osowski, S.; Hoai, L.T.; Markiewicz, T. Support vector machine based expert system for reliable heartbeat recognition. IEEE Trans. Biomed. Eng. 2004, 51, 582–589. [Google Scholar] [CrossRef] [PubMed]
- Willems, J.L.; Lesaffre, E. Comparison of multigroup logistic and linear discriminant ecg and vcg classification. J. Electrocardiol. 1987, 20, 83–92. [Google Scholar] [CrossRef]
- Hu, Y.H.; Tompkins, W.J.; Urrusti, J.L.; Afonso, V.X. Applications of artificial neural networks for ECG signal detection and classification. J. Electrocardiol. 1993, 26, 66–73. [Google Scholar] [PubMed]
- Trahanias, P.; Skordalakis, E. Syntactic pattern recognition of the ECG. IEEE Trans. Pattern Anal. Mach. Intell. 1990, 12, 648–657. [Google Scholar] [CrossRef] [Green Version]
- Inan, O.T.; Giovangrandi, L.; Kovacs, G.T. Robust neural-network-based classification of premature ventricular contractions using wavelet transform and timing interval features. IEEE Trans. Biomed. Eng. 2006, 53, 2507–2515. [Google Scholar] [CrossRef]
- Hu, Y.H.; Palreddy, S.; Tompkins, W.J. A patient-adaptable ECG beat classifier using a mixture of experts approach. IEEE Trans. Biomed. Eng. 1997, 44, 891–900. [Google Scholar]
- Dehan, L.; Guanggui, X.U.; Yuhua, Z.; Hosseini, H.G. Novel ECG diagnosis model based on multi-stage artificial neural networks. Chin. J. Sci. Instrum. 2008, 29, 27. [Google Scholar]
- Ceylan, R.; Ozbay, Y. Comparison of FCM, PCA and WT techniques for classification ECG arrhythmias using artificial neural network. Expert Syst. Appl. 2007, 33, 286–295. [Google Scholar] [CrossRef]
- Polat, K.; Günes, S. Breast cancer diagnosis using least square support vector machine. Digit. Signal Process. 2007, 17, 694–701. [Google Scholar] [CrossRef]
- Dreiseitl, S.; Ohno-Machado, L.; Kittler, H.; Vinterbo, S.; Billhardt, H.; Binder, M. A comparison of machine learning methods for the diagnosis of pigmented skin lesions. J. Biomed. Inform. 2001, 34, 28–36. [Google Scholar] [CrossRef] [Green Version]
- Shafiq, M.; Yu, X.; Bashir, A.K.; Chaudhry, H.N.; Wang, D. A machine learning approach for feature selection traffic classification using security analysis. J. Supercomput. 2018, 74, 4867–4892. [Google Scholar] [CrossRef]
- Bashir, A.K.; Arul, R.; Basheer, S.; Raja, G.; Jayaraman, R.; Qureshi, N.M.F. An optimal multitier resource allocation of cloud RAN in 5G using machine learning. Trans. Emerg. Telecommun. Technol. 2019, 30, 3627. [Google Scholar] [CrossRef]
- Kononenko, I. Machine learning for medical diagnosis: History, state of the art and perspective. Artif. Intell. Med. 2001, 23, 89–109. [Google Scholar] [CrossRef]
- Ecar, A. Recommended practice for testing and reporting performance results of ventricular arrhythmia detection algorithms. Assoc. Adv. Med. Instrum. 1987, 69. [Google Scholar]
- Huertas-Fernandez, I.; Garcia-Gomez, F.J.; Garcia-Solis, D.; Benitez-Rivero, S.; Marin-Oyaga, V.A.; Jesus, S.; Mir, P. Machine learning models for the differential diagnosis of vascular parkinsonism and Parkinson’s disease using [123 I] FP-CIT SPECT. Eur. J. Nucl. Med. Mol. Imaging 2015, 42, 112–119. [Google Scholar] [CrossRef] [PubMed]
- Salvatore, C.; Cerasa, A.; Battista, P.; Gilardi, M.C.; Quattrone, A.; Castiglioni, I. Magnetic resonance imaging biomarkers for the early diagnosis of Alzheimer’s disease: A machine learning approach. Front. Neurosci. 2015, 9, 307. [Google Scholar] [CrossRef] [Green Version]
- Kiranyaz, S.; Ince, T.; Gabbouj, M. Real-time patient-specific ECG classification by 1-D convolutional neural networks. IEEE Trans. Biomed. Eng. 2015, 63, 664–675. [Google Scholar] [CrossRef]
- Rajpurkar, P.; Hannun, A.Y.; Haghpanahi, M.; Bourn, C.; Ng, A.Y. Cardiologist-level arrhythmia detection with convolutional neural networks. arXiv 2017, arXiv:1707.01836. [Google Scholar]
- Acharya, U.R.; Oh, S.L.; Hagiwara, Y.; Tan, J.H.; Adam, M.; Gertych, A.; San Tan, R. A deep convolutional neural network model to classify heartbeats. Comput. Biol. Med. 2017, 89, 389–396. [Google Scholar] [CrossRef]
- Chen, J.; Valehi, A.; Razi, A. Smart Heart Monitoring: Early Prediction of Heart Problems Through Predictive Analysis of ECG Signals. IEEE Access 2019, 7, 120831–120839. [Google Scholar] [CrossRef]
- Lee, S.C. Using a translation-invariant neural network to diagnose heart arrhythmia. In Advances in Neural Information Processing Systems; Morgan Kaufmann: San Francisco, CA, USA, 1990; pp. 240–247. [Google Scholar]
- De Chazal, P.; Reilly, R.B. A patient-adapting heartbeat classifier using ECG morphology and heartbeat interval features. IEEE Trans. Biomed. Eng. 2015, 53, 2535–2543. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Z.; Stiles, M.K.; Zhao, J. Robust ECG signal classification for detection of atrial fibrillation using a novel neural network. In Proceedings of the 2017 Computing in Cardiology (CinC), Rennes, France, 24–27 September 2017; IEEE: Piscataway, NJ, USA, 2017; pp. 1–4. [Google Scholar]
- Clevert, D.A.; Unterthiner, T.; Hochreiter, S. Fast and accurate deep network learning by exponential linear units (elus). arXiv 2015, arXiv:1511.07289. [Google Scholar]
- Li, D.; Zhang, J.; Zhang, Q.; Wei, X. Classification of ECG signals based on 1D convolution neural network. In Proceedings of the 2017 IEEE 19th International Conference on e-Health Networking, Applications and Services (Healthcom), Dalian, China, 12–15 October 2017; IEEE: Piscataway, NJ, USA, 2017; pp. 1–6. [Google Scholar]
- Jun, T.J.; Nguyen, H.M.; Kang, D.; Kim, D.; Kim, D.; Kim, Y.H. ECG arrhythmia classification using a 2-D convolutional neural network. arXiv 2018, arXiv:1804.06812. [Google Scholar]
- Mohanty, M.D.; Mohanty, B.; Mohanty, M.N. R-peak detection using efficient technique for tachycardia detection. In Proceedings of the 2017 2nd International Conference on Man and Machine Interfacing (MAMI), Bhubaneswar, India, 21–23 December 2017; IEEE: Piscataway, NJ, USA, 2017; pp. 1–5. [Google Scholar]
- Moody, G.B.; Mark, R.G. The impact of the MIT-BIH arrhythmia database. IEEE Eng. Med. Biol. Mag. 2001, 20, 45–50. [Google Scholar] [CrossRef] [PubMed]
- Abadi, M.; Agarwal, A.; Barham, P.; Brevdo, E.; Chen, Z.; Citro, C.; Corrado, G.S.; Davis, A.; Dean, J.; Devin, M.; et al. Tensorflow: Large-scale machine learning on heterogeneous distributed systems. arXiv 2016, arXiv:1603.04467. [Google Scholar]
- Übeyli, E.D. Combining recurrent neural networks with eigenvector methods for classification of ECG beats. Digit. Signal Process. 2009, 19, 320–329. [Google Scholar] [CrossRef]
- Dutta, S.; Chatterjee, A.; Munshi, S. Correlation technique and least square support vector machine combine for frequency domain based ECG beat classification. Med. Eng. Phys. 2010, 32, 1161–1169. [Google Scholar] [CrossRef]
- Kumar, R.G.; Kumaraswamy, Y.S. Investigating cardiac arrhythmia in ECG using random forest classification. Int. J. Comput. Appl. 2012, 37, 31–34. [Google Scholar]
- Park, J.; Lee, K.; Kang, K. Arrhythmia detection from heartbeat using k-nearest neighbor classifier. In Proceedings of the 2013 IEEE International Conference on Bioinformatics and Biomedicine, Shanghai, China, 18–21 December 2013; IEEE: Piscataway, NJ, USA, 2013; pp. 15–22. [Google Scholar]
- Ince, T.; Kiranyaz, S.; Eren, L.; Askar, M.; Gabbouj, M. Real-time motor fault detection by 1-D convolutional neural networks. IEEE Trans. Ind. Electron. 2016, 63, 7067–7075. [Google Scholar] [CrossRef]
- Izci, E.; Ozdemir, M.A.; Degirmenci, M.; Akan, A. Cardiac Arrhythmia Detection from 2D ECG Images by Using Deep Learning Technique. In Proceedings of the 2019 Medical Technologies Congress (TIPTEKNO), Selçuk, Turkey, 3–5 October 2019; IEEE: Piscataway, NJ, USA, 2019; pp. 1–4. [Google Scholar]
- Rajkumar, A.; Ganesan, M.; Lavanya, R. Arrhythmia classification on ECG using Deep Learning. In Proceedings of the 2019 5th International Conference on Advanced Computing and Communication Systems (ICACCS), Coimbatore, India, 15–16 March 2019; IEEE: Piscataway, NJ, USA, 2019; pp. 365–369. [Google Scholar]
- Guler, I.; Ubeylı, E.D. ECG beat classifier designed by combined neural network model. Pattern Recognit. 2005, 38, 199–208. [Google Scholar] [CrossRef]
- Yu, S.N.; Chou, K.T. Integration of independent component analysis and neural networks for ECG beat classification. Expert Syst. Appl. 2008, 34, 2841–2846. [Google Scholar] [CrossRef]
- Melgani, F.; Bazi, Y. Classification of electrocardiogram signals with support vector machines and particle swarm optimization. IEEE Trans. Inf. Technol. Biomed. 2008, 12, 667–677. [Google Scholar] [CrossRef] [PubMed]





| Layers | Type | Filter Size | Stride | Kernel | Input Size | Parameters |
|---|---|---|---|---|---|---|
| Layer 1 | Conv2-D | 3 × 3 | 1 | 64 | 256 × 256 × 1 | 576 |
| Layer 2 | Pooling | 2 × 2 | 2 | - | 256 × 256 × 64 | - |
| Layer 3 | Conv2-D | 3 × 3 | 1 | 128 | 128 × 128 × 64 | 73,728 |
| Layer 4 | Pooling | 2 × 2 | 2 | - | 128 × 128 × 128 | - |
| Layer 5 | Conv2-D | 3 × 3 | 1 | 256 | 64 × 64 × 128 | 294,912 |
| Layer 6 | Pooling | 2 × 2 | 2 | - | 64 × 64 × 256 | - |
| Layer 7 | Conv2-D | 3 × 3 | 1 | 512 | 32 × 32 × 256 | 1,179,648 |
| Layer 8 | Pooling | 2 × 2 | 2 | - | 32 × 32 × 512 | - |
| Layer 9 | Fully Connected | - | - | 4096 | 16 × 16 × 512 | 2,097,152 |
| Layer 10 | Output Layer | - | - | 8 | 4096 | 32,776 |
| Learning Rate | Batch Size | Average Accuracy |
|---|---|---|
| 0.001 | 2800 | 99.11 |
| 0.001 | 2000 | 98.96 |
| 0.001 | 1000 | 99.00 |
| 0.001 | 500 | 98.95 |
| 0.001 | 100 | 98.93 |
| Batch Size | Learning Rate | Average Accuracy |
|---|---|---|
| 2800 | 0.001 | 99.11 |
| 2800 | 0.005 | 98.84 |
| 2800 | 0.100 | 98.89 |
| 2800 | 0.200 | 98.91 |
| Model | Native/Augmentation | Classes | Accuracy % | Sensitivity % | Specificity % | Precision % | F1 Score |
|---|---|---|---|---|---|---|---|
| FFNN [59] | 4 | 96.94 | 96.31 | 97.78 | - | - | |
| PNN [60] | 8 | 98.71 | - | 99.65 | - | - | |
| SVM [61] | 6 | 91.67 | 93.83 | 90.49 | - | - | |
| RNN [52] | 4 | 98.06 | 98.15 | 97.78 | - | - | |
| LS-SVM [53] | 3 | 95.82 | 86.16 | 99.17 | 97.01 | 0.91 | |
| RFT [54] | 3 | 92.16 | - | - | - | - | |
| KNN [55] | 17 | 97.00 | 96.60 | 95.80 | - | - | |
| 1-D CNN [56] | 5 | 96.40 | 68.80 | 99.50 | 79.20 | 0.73 | |
| AlexNet [48] | Augmented | 8 | 98.85 | 97.08 | 99.62 | 98.59 | 0.97 |
| AlexNet [48] | Native | 8 | 98.81 | 96.81 | 99.68 | 98.63 | 0.97 |
| VGGNet [48] | Augmented | 8 | 98.63 | 96.93 | 99.37 | 97.86 | 0.97 |
| VGGNet [48] | Native | 8 | 98.77 | 97.26 | 99.43 | 98.08 | 0.97 |
| 2-D CNN [57] | 5 | 97.42 | - | - | - | ||
| 1-D CNN [58] | 7 | 93.60 | - | - | - | ||
| Proposed (1-D) | Native | 8 | 97.80 | - | - | - | - |
| Proposed (2-D) | Augmented | 8 | 99.11 | 97.91 | 99.61 | 98.58 | 0.98 |
| Proposed (2-D) | Native | 8 | 98.92 | 97.26 | 99.67 | 98.69 | 0.98 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ullah, A.; Anwar, S.M.; Bilal, M.; Mehmood, R.M. Classification of Arrhythmia by Using Deep Learning with 2-D ECG Spectral Image Representation. Remote Sens. 2020, 12, 1685. https://0-doi-org.brum.beds.ac.uk/10.3390/rs12101685
Ullah A, Anwar SM, Bilal M, Mehmood RM. Classification of Arrhythmia by Using Deep Learning with 2-D ECG Spectral Image Representation. Remote Sensing. 2020; 12(10):1685. https://0-doi-org.brum.beds.ac.uk/10.3390/rs12101685
Chicago/Turabian StyleUllah, Amin, Syed Muhammad Anwar, Muhammad Bilal, and Raja Majid Mehmood. 2020. "Classification of Arrhythmia by Using Deep Learning with 2-D ECG Spectral Image Representation" Remote Sensing 12, no. 10: 1685. https://0-doi-org.brum.beds.ac.uk/10.3390/rs12101685