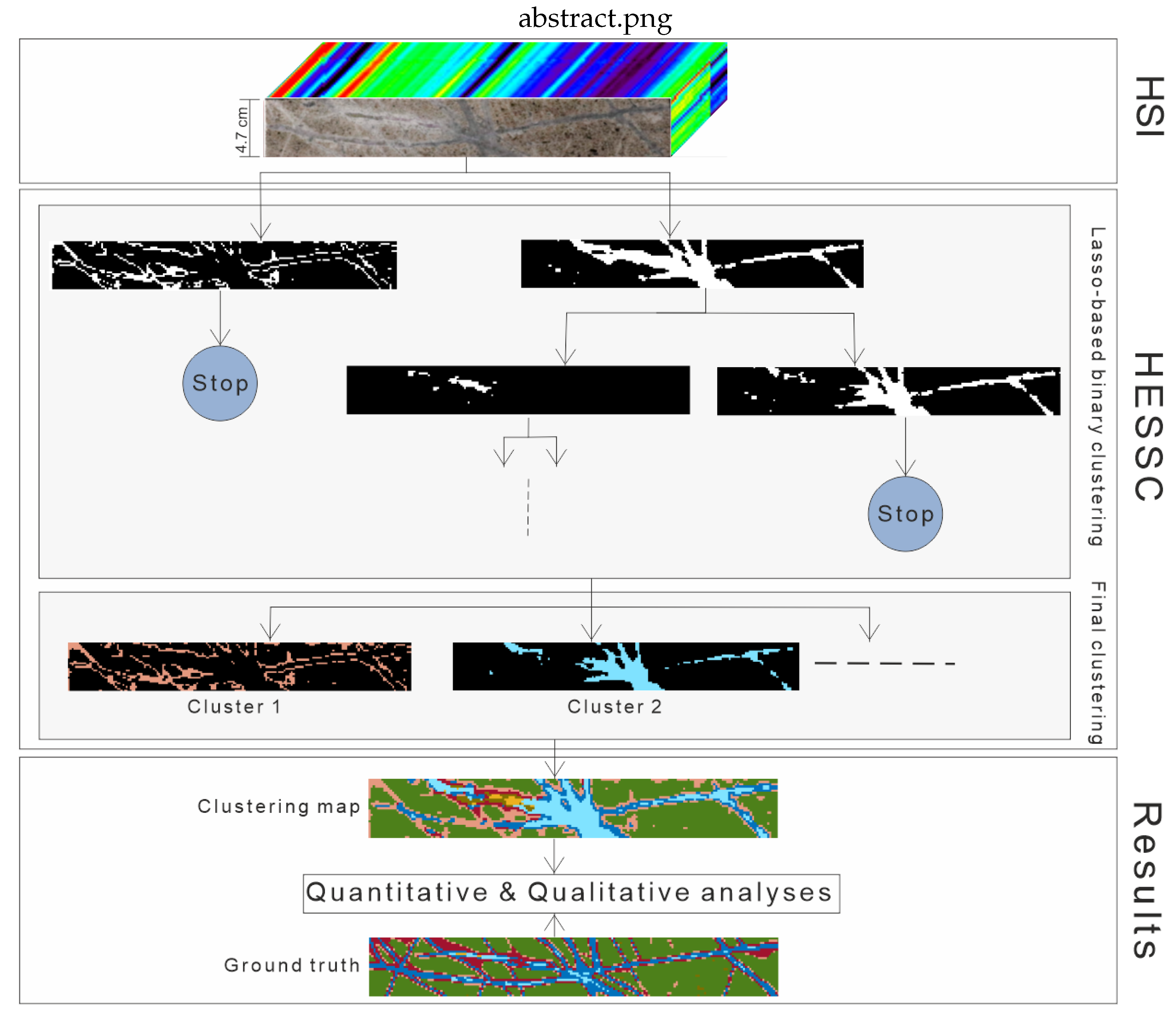
Hierarchical Sparse Subspace Clustering (HESSC): An Automatic Approach for Hyperspectral Image Analysis
Abstract
:1. Introduction
- We propose a sparse subspace-based clustering algorithm that considerably reduces the computational power requirements by using a novel lasso-binary clustering algorithm.
- The proposed algorithm is able to provide robust and stable results thanks to the incorporation of the entropy-based consensus algorithm within its structure, which decreases the effect of the initial random selection of a sample in the lasso-binary clustering algorithm.
- The proposed algorithm benefits from defined criteria, which make it possible to automatically generate the number of clusters.
2. Methodology
2.1. Overview on the State-of-the-Art Sparse Subspace-Based Clustering Algorithm
2.2. Hierarchical Sparse Subspace Clustering (HESSC)
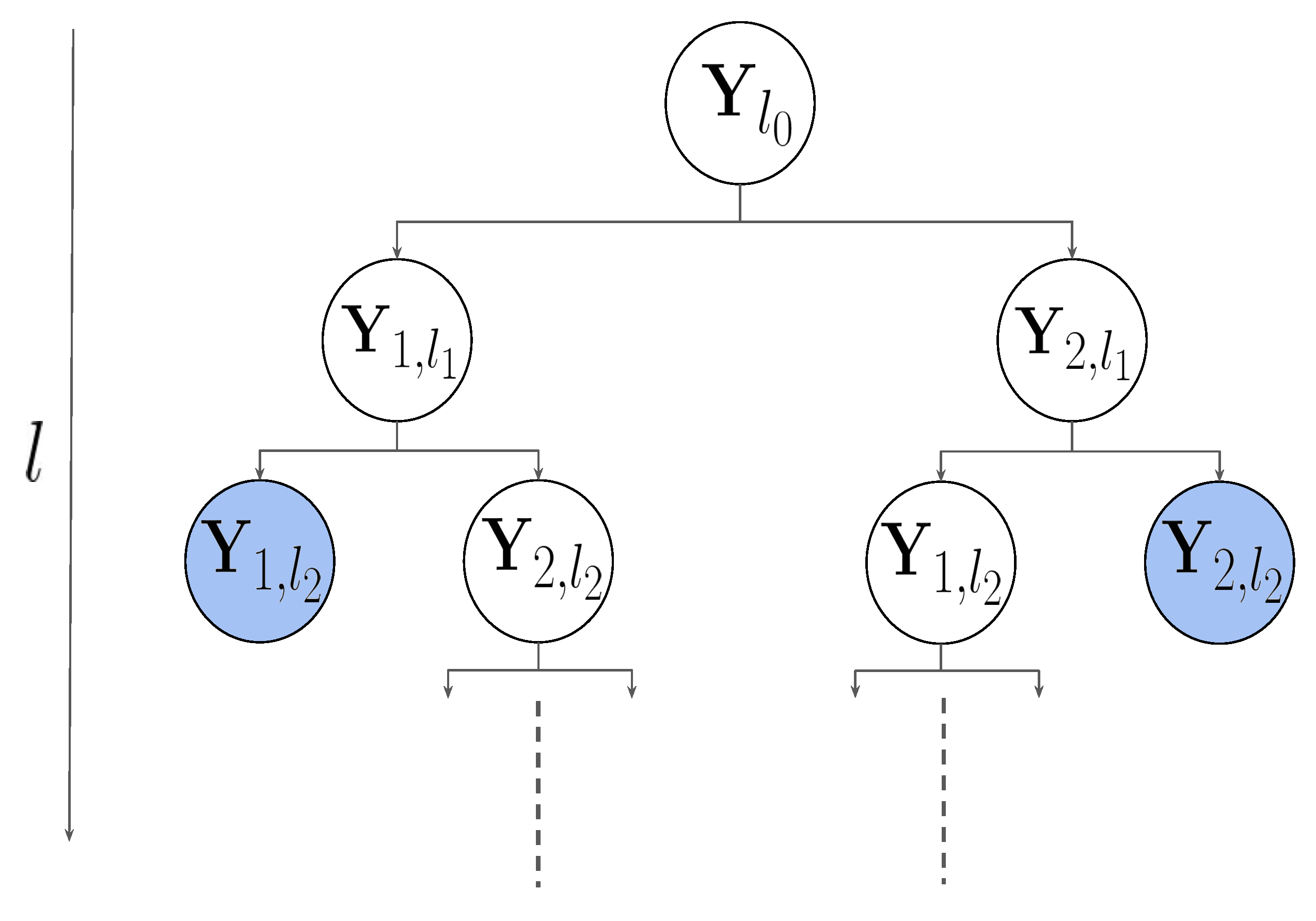
2.2.1. Hierarchical Structure in HESSC
2.2.2. Binary Clustering in HESSC
2.2.3. Automatic Generation of Number of Clusters
3. Experimental Results
3.1. Data Acquisition and Description
3.2. Evaluation Metrics
3.3. Quantitative Analyses on Clustering Results
3.4. Execution Time
3.5. Qualitative Analyses on Clustering Results
3.6. Evaluation of the Automatically-Generated Number of Clusters
3.7. Sensitivity of Parameters in HESSC Algorithm
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| HSIs | Hyperspectral images |
| SSC | Sparse subspace clustering |
| SSC-OMP | Sparse subspace clustering by orthogonal matching pursuit |
| ESC | Exemplar-based subspace clustering |
| ECC | Entropy-based consensus clustering |
| HESSC | Hierarchical sparse subspace-based clustering |
| CA | Clustering accuracy |
| ARI | Adjust rand index |
References
- Goetz, A.F.; Vane, G.; Solomon, J.E.; Rock, B.N. Imaging Spectrometry for Earth Remote Sensing. Science 1985, 228, 1147–1153. [Google Scholar] [CrossRef]
- Tarabalka, Y.; Chanussot, J.; Benediktsson, J.A. Segmentation and classification of hyperspectral images using minimum spanning forest grown from automatically selected markers. IEEE Trans. Syst. Man Cybern. Part B 2009, 40, 1267–1279. [Google Scholar] [CrossRef] [Green Version]
- Ghamisi, P.; Maggiori, E.; Li, S.; Souza, R.; Tarablaka, Y.; Moser, G.; De Giorgi, A.; Fang, L.; Chen, Y.; Chi, M.; et al. New Frontiers in Spectral-Spatial Hyperspectral Image Classification: The Latest Advances Based on Mathematical Morphology, Markov Random Fields, Segmentation, Sparse Representation, and Deep Learning. IEEE Geosci. Remote Sens. Mag. 2018, 6, 10–43. [Google Scholar] [CrossRef]
- Rasti, B.; Hong, D.; Hang, R.; Ghamisi, P.; Kang, X.; Chanussot, J.; Benediktsson, J.A. Feature Extraction for Hyperspectral Imagery: The Evolution from Shallow to Deep (Overview and Toolbox). IEEE Geosci. Remote Sens. Mag. 2020. [Google Scholar] [CrossRef]
- Lorenz, S.; Seidel, P.; Ghamisi, P.; Zimmermann, R.; Tusa, L.; Khodadadzadeh, M.; Contreras, I.C.; Gloaguen, R. Multi-Sensor Spectral Imaging of Geological Samples: A Data Fusion Approach Using Spatio-Spectral Feature Extraction. Sensors 2019, 19, 2787. [Google Scholar] [CrossRef] [Green Version]
- Acosta, I.C.C.; Khodadadzadeh, M.; Tusa, L.; Ghamisi, P.; Gloaguen, R. A Machine Learning Framework for Drill-Core Mineral Mapping Using Hyperspectral and High-Resolution Mineralogical Data Fusion. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2019, 12, 4829–4842. [Google Scholar] [CrossRef]
- Tusa, L.; Andreani, L.; Khodadadzadeh, M.; Contreras, C.; Ivascanu, P.; Gloaguen, R.; Gutzmer, J. Mineral Mapping and Vein Detection in Hyperspectral Drill-Core Scans: Application to Porphyry-Type Mineralization. Minerals 2019, 9, 122. [Google Scholar] [CrossRef] [Green Version]
- Hughes, G. On the mean accuracy of statistical pattern recognizers. IEEE Trans. Inf. Theory 1968, IT, 55–63. [Google Scholar] [CrossRef] [Green Version]
- Ghamisi, P.; Yokoya, N.; Li, J.; Liao, W.; Liu, S.; Plaza, J.; Rasti, B.; Plaza, A. Advances in Hyperspectral Image and Signal Processing: A Comprehensive Overview of the State of the Art. IEEE Geosci. Remote Sens. Mag. 2017, 5, 37–78. [Google Scholar] [CrossRef] [Green Version]
- Benediktsson, J.A.; Palmason, J.A.; Sveinsson, J.R. Classification of hyperspectral data from urban areas based on extended morphological profiles. IEEE Trans. Geosci. Remote Sens. 2005, 43, 480–491. [Google Scholar] [CrossRef]
- Adão, T.; Hruška, J.; Pádua, L.; Bessa, J.; Peres, E.; Morais, R.; Sousa, J.J. Hyperspectral Imaging: A Review on UAV-Based Sensors, Data Processing and Applications for Agriculture and Forestry. Remote Sens. 2017, 9, 1110. [Google Scholar] [CrossRef] [Green Version]
- Ghamisi, P.; Plaza, J.; Chen, Y.; Li, J.; Plaza, A.J. Advanced Spectral Classifiers for Hyperspectral Images: A review. IEEE Geosci. Remote Sens. Mag. 2017, 5, 8–32. [Google Scholar] [CrossRef] [Green Version]
- Saxena, A.; Prasad, M.; Gupta, A.; Bharill, N.; Patel, O.P.; Tiwari, A.; Er, M.J.; Ding, W.; Lin, C.T. A review of clustering techniques and developments. Neurocomputing 2017, 267, 664–681. [Google Scholar] [CrossRef] [Green Version]
- Yang, W.; Hou, K.; Liu, B.; Yu, F.; Lin, L. Two-Stage Clustering Technique Based on the Neighboring Union Histogram for Hyperspectral Remote Sensing Images. IEEE Access 2017, 5, 5640–5647. [Google Scholar] [CrossRef]
- Elhamifar, E.; Vidal, R. Sparse Subspace Clustering: Algorithm, Theory, and Applications. IEEE Trans. Pattern Anal. Mach. Intell. 2013, 35, 2765–2781. [Google Scholar] [CrossRef] [Green Version]
- Olshausen, B.A. Emergence of simple-cell receptive field properties by learning a sparse code for natural images. Nature 1996, 381, 607–609. [Google Scholar] [CrossRef]
- Wright, J.; Yang, A.Y.; Ganesh, A.; Sastry, S.S.; Ma, Y. Robust Face Recognition via Sparse Representation. IEEE Trans. Pattern Anal. Mach. Intell. 2009, 31, 210–227. [Google Scholar] [CrossRef] [Green Version]
- Wright, J.; Ma, Y.; Mairal, J.; Sapiro, G.; Huang, T.S.; Yan, S. Sparse Representation for Computer Vision and Pattern Recognition. Proc. IEEE 2010, 98, 1031–1044. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; Nasrabadi, N.M.; Tran, T.D. Hyperspectral Image Classification Using Dictionary-Based Sparse Representation. IEEE Trans. Geosci. Remote Sens. 2011, 49, 3973–3985. [Google Scholar] [CrossRef]
- Srinivas, U.; Chen, Y.; Monga, V.; Nasrabadi, N.M.; Tran, T.D. Exploiting Sparsity in Hyperspectral Image Classification via Graphical Models. IEEE Geosci. Remote Sens. Lett. 2013, 10, 505–509. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; Nasrabadi, N.M.; Tran, T.D. Hyperspectral Image Classification via Kernel Sparse Representation. IEEE Trans. Geosci. Remote Sens. 2013, 51, 217–231. [Google Scholar] [CrossRef] [Green Version]
- Fang, L.; Li, S.; Kang, X.; Benediktsson, J.A. Spatial Classification of Hyperspectral Images with a Superpixel-Based Discriminative Sparse Model. IEEE Trans. Geosci. Remote Sens. 2015, 53, 4186–4201. [Google Scholar] [CrossRef]
- Fang, L.; Li, S.; Kang, X.; Benediktsson, J.A. Spatial Hyperspectral Image Classification via Multiscale Adaptive Sparse Representation. IEEE Trans. Geosci. Remote Sens. 2014, 52, 7738–7749. [Google Scholar] [CrossRef]
- Fu, W.; Li, S.; Fang, L.; Kang, X.; Benediktsson, J.A. Hyperspectral Image Classification Via Shape-Adaptive Joint Sparse Representation. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2016, 9, 556–567. [Google Scholar] [CrossRef]
- Li, J.; Zhang, H.; Zhang, L. Efficient Superpixel-Level Multitask Joint Sparse Representation for Hyperspectral Image Classification. IEEE Trans. Geosci. Remote Sens. 2015, 53, 5338–5351. [Google Scholar] [CrossRef]
- You, C.; Li, C.; Robinson, D.P.; Vidal, R. Scalable Exemplar-based Subspace Clustering on Class-Imbalanced Data. In Proceedings of the European Conference on Computer Vision (ECCV), Munich, Germany, 8–14 September 2018. [Google Scholar]
- Shahi, K.R.; Khodadadzadeh, M.; Tolosana-delgado, R.; Tusa, L.; Gloaguen, R. The Application of Subspace Clustering Algorithms in Drill-Core Hyperspectral Domaining. In Proceedings of the 2019 10th Workshop on Hyperspectral Imaging and Signal Processing: Evolution in Remote Sensing (WHISPERS), Amsterdam, The Netherlands, 24–26 September 2019; pp. 1–5. [Google Scholar] [CrossRef]
- Zhang, H.; Zhai, H.; Zhang, L.; Li, P. Spectral–Spatial Sparse Subspace Clustering for Hyperspectral Remote Sensing Images. IEEE Trans. Geosci. Remote Sens. 2016, 54, 3672–3684. [Google Scholar] [CrossRef]
- Hinojosa, C.; Bacca, J.; Arguello, H. Coded Aperture Design for Compressive Spectral Subspace Clustering. IEEE J. Sel. Top. Signal Process. 2018, 12, 1589–1600. [Google Scholar] [CrossRef]
- Dyer, E.; Sankaranarayanan, A.; Baraniuk, R. Greedy feature selection for subspace clustering. J. Mach. Learn. Res. 2013, 14, 2487–2517. [Google Scholar]
- You, C.; Robinson, D.; Vidal, R. Scalable sparse subspace clustering by orthogonal matching pursuit. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 3918–3927. [Google Scholar]
- Wu, T.; Gurram, P.; Rao, R.M.; Bajwa, W.U. Hierarchical union-of-subspaces model for human activity summarization. In Proceedings of the IEEE International Conference on Computer Vision Workshops, Santiago, Chile, 11–18 December 2015; pp. 1–9. [Google Scholar]
- Liu, H.; Zhao, R.; Fang, H.; Cheng, F.; Fu, Y.; Liu, Y.Y. Entropy-based consensus clustering for patient stratification. Bioinformatics 2017, 33, 2691–2698. [Google Scholar] [CrossRef]
- Iordache, M.; Bioucas-Dias, J.M.; Plaza, A. Sparse Unmixing of Hyperspectral Data. IEEE Trans. Geosci. Remote Sens. 2011, 49, 2014–2039. [Google Scholar] [CrossRef] [Green Version]
- Ifarraguerri, A.; Chang, C. Multispectral and hyperspectral image analysis with convex cones. IEEE Trans. Geosci. Remote Sens. 1999, 37, 756–770. [Google Scholar] [CrossRef] [Green Version]
- Balzano, L.; Nowak, R.; Bajwa, W. Column subset selection with missing data. In Proceedings of the NIPS Workshop on Low-Rank Methods for Large-Scale Machine Learning, Whistler, BC, Canada, 11 December 2010; Volume 1. [Google Scholar]
- Esser, E.; Moller, M.; Osher, S.; Sapiro, G.; Xin, J. A convex model for nonnegative matrix factorization and dimensionality reduction on physical space. IEEE Trans. Image Process. 2012, 21, 3239–3252. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Elhamifar, E.; Sapiro, G.; Vidal, R. See all by looking at a few: Sparse modeling for finding representative objects. In Proceedings of the 2012 IEEE Conference on Computer Vision and Pattern Recognition, Providence, RI, USA, 16–21 June 2012; pp. 1600–1607. [Google Scholar] [CrossRef] [Green Version]
- Boyd, S.; Parikh, N.; Chu, E.; Peleato, B.; Eckstein, J. Distributed Optimization and Statistical Learning via the Alternating Direction Method of Multipliers; Now Publishers Inc.: Delft, The Netherlands, 2011; pp. 1–122. [Google Scholar]
- Benzi, M.; Olshanskii, M.A. An augmented Lagrangian-based approach to the Oseen problem. SIAM J. Sci. Comput. 2006, 28, 2095–2113. [Google Scholar] [CrossRef]
- Efron, B.; Hastie, T.; Johnstone, I.; Tibshirani, R. Least angle regression. Ann. Stat. 2004, 32, 407–499. [Google Scholar] [CrossRef] [Green Version]
- Mairal, J.; Bach, F.; Ponce, J.; Sapiro, G. Online Learning for Matrix Factorization and Sparse Coding. J. Mach. Learn. Res. 2009, 11. [Google Scholar] [CrossRef]
- Vedaldi, A.; Fulkerson, B. VLFeat: An open and portable library of computer vision algorithms. In Proceedings of the 18th ACM International Conference on Multimedia, Florence, Italy, 25–29 October 2010; Volume 3, pp. 1469–1472. [Google Scholar] [CrossRef]
- Wu, J.; Xiong, H.; Chen, J. Adapting the Right Measures for K-means Clustering. In Proceedings of the 15th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, Paris, France, 28 June–1 July 2009; pp. 877–886. [Google Scholar] [CrossRef]
- Wagner, S.; Wagner, D. Comparing Clusterings: An Overview; Universität Karlsruhe, Fakultät für Informatik Karlsruhe: Karlsruhe, Germany, 2007. [Google Scholar]
- Hubert, L.; Arabie, P. Comparing partitions. J. Classif. 1985, 2, 193–218. [Google Scholar] [CrossRef]
- Rand, W.M. Objective Criteria for the Evaluation of Clustering Methods. J. Am. Stat. Assoc. 1971, 66, 846–850. [Google Scholar] [CrossRef]





| Evaluation Metric | K-Means | FCM | ECC | SSC | SSC-OMP | ESC | HESSC |
|---|---|---|---|---|---|---|---|
| CA | 33.08 ± 2.41 | 27.80 ± 0.00 | 28.36 ± 0.72 | 28.98 ± 0.00 | 45.39 ± 0.83 | 30.47 ± 5.17 | 53.12 ± 0.00 |
| F-measure | 20.04 ± 2.41 | 18.87 ± 0.00 | 18.97 ± 0.25 | 18.81 ± 0.00 | 22.52 ± 0.11 | 18.37 ± 2.55 | 23.21 ± 0.00 |
| ARI | 12.67 ± 1.60 | 9.13 ± 0.00 | 8.82 ± 0.93 | 10.53 ± 0.00 | 16.18 ± 0.67 | 9.10 ± 3.88 | 31.02 ± 0.00 |
| Evaluation Metric | K-Means | FCM | ECC | SSC | SSC-OMP | ESC | HESSC |
|---|---|---|---|---|---|---|---|
| CA | 36.89 ± 3.5 | 30.59 ± 0.82 | 39.38 ± 1.68 | 27.72 ± 0.13 | 36.55 ± 2.10 | 43.49 ± 11.19 | 46.53 ± 0.00 |
| F-measure | 22.61 ± 1.57 | 20.20 ± 0.36 | 23.44 ± 0.59 | 18.23 ± 0.00 | 19.68 ± 0.16 | 26.26 ± 5.85 | 22.23 ± 0.00 |
| ARI | 18.68 ± 2.38 | 15.09 ± 0.64 | 20.60 ± 1.50 | 15.23 ± 0.00 | 17.19 ± 0.69 | 22.07 ± 8.99 | 22.09 ± 0.00 |
| Evaluation Metric | K-Means | FCM | ECC | SSC | SSC-OMP | ESC | HESSC |
|---|---|---|---|---|---|---|---|
| CA | 49.02 ± 0.00 | 38.93 ± 0.00 | 49.02 ± 0.00 | 33.31 ± 0.00 | 40.11 ± 0.00 | 50.03 ± 11.19 | 61.31 ± 0.58 |
| F-measure | 32.20 ± 0.00 | 30.10 ± 0.00 | 32.16 ± 0.00 | 23.72 ± 0.00 | 24.71 ± 0.00 | 27.83 ± 3.14 | 32.69 ± 0.44 |
| ARI | 11.42 ± 0.00 | 8.60 ± 0.00 | 11.30 ± 0.00 | 4.06 ± 0.00 | -0.71 ± 0.00 | 9.64 ± 4.72 | 12.94 ± 0.69 |
| Evaluation Metric | K-Means | FCM | ECC | SSC | SSC-OMP | ESC | HESSC |
|---|---|---|---|---|---|---|---|
| CA | 49.38 ± 0.00 | 53.88 ± 0.00 | 51.40 ± 0.00 | - | 34.80 ± 0.00 | 33.66 ± 0.02 | 56.39 ± 0.00 |
| F-measure | 45.04 ± 0.00 | 45.50 ± 0.00 | 40.79 ± 0.00 | - | 22.02 ± 0.00 | 29.36 ± 0.00 | 46.99 ± 0.00 |
| ARI | 26.63 ± 0.00 | 28.44 ± 0.00 | 35.43 ± 0.00 | - | 7.74 ± 0.00 | 20.78 ± 0.00 | 36.55 ± 0.00 |
| Sample No. | K-Means | FCM | ECC | SSC | SSC-OMP | ESC | HESSC |
|---|---|---|---|---|---|---|---|
| Sample No. 1 | 0.46 ± 0.13 | 9.14 ± 1.12 | 43.50 ± 18.33 | 235.58 ± 24.57 | 1.00 ± 0.08 | 4.46 ± 0.15 | 38.46 ± 1.47 |
| Sample No. 2 | 0.60 ± 0.19 | 9.48 ± 0.49 | 44.95 ± 1.98 | 319.38 ± 91.17 | 1.01 ± 0.07 | 9.41 ± 0.47 | 34.62 ± 1.16 |
| Sample No. 3 | 0.67 ± 0.34 | 5.41 ± 0.15 | 37.14 ± 2.52 | 363.81 ± 34.04 | 1.10 ± 0.13 | 10.55 ± 0.66 | 48.83 ± 1.32 |
| Trento | 2.55 ± 0.00 | 24.38 ± 0.00 | 162.52 ± 0.00 | - | 518.27 ± 1.06 | 259.54 ± 0.00 | 478.94 ± 0.00 |
| 0.1 | 0.2 | 0.3 | 0.4 | 0.5 | 0.6 | 0.7 | 0.8 | 0.9 | |
| No. Clusters | 2 | 2 | 4 | 4 | 4 | 5 | 4 | 4 | 2 |
| 0 | 0.1 | 0.2 | 0.3 | 0.4 | 0.5 | 0.6 | 0.7 | 0.8 | 0.9 | 1 | |
| No. Clusters | 8 | 6 | 7 | 7 | 4 | 4 | 3 | 2 | 2 | 2 | 2 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rafiezadeh Shahi, K.; Khodadadzadeh, M.; Tusa, L.; Ghamisi, P.; Tolosana-Delgado, R.; Gloaguen, R. Hierarchical Sparse Subspace Clustering (HESSC): An Automatic Approach for Hyperspectral Image Analysis. Remote Sens. 2020, 12, 2421. https://0-doi-org.brum.beds.ac.uk/10.3390/rs12152421
Rafiezadeh Shahi K, Khodadadzadeh M, Tusa L, Ghamisi P, Tolosana-Delgado R, Gloaguen R. Hierarchical Sparse Subspace Clustering (HESSC): An Automatic Approach for Hyperspectral Image Analysis. Remote Sensing. 2020; 12(15):2421. https://0-doi-org.brum.beds.ac.uk/10.3390/rs12152421
Chicago/Turabian StyleRafiezadeh Shahi, Kasra, Mahdi Khodadadzadeh, Laura Tusa, Pedram Ghamisi, Raimon Tolosana-Delgado, and Richard Gloaguen. 2020. "Hierarchical Sparse Subspace Clustering (HESSC): An Automatic Approach for Hyperspectral Image Analysis" Remote Sensing 12, no. 15: 2421. https://0-doi-org.brum.beds.ac.uk/10.3390/rs12152421








