Characterizing Land Surface Phenology and Exotic Annual Grasses in Dryland Ecosystems Using Landsat and Sentinel-2 Data in Harmony
Abstract
:1. Introduction
2. Material and Methods
2.1. Automated Cloud, Shadow, Snow/Ice, and Water Masking Procedure
2.2. Automated Time-Series Modeling and Metrics
2.3. In Situ Observations of Exotic Annual Grass Cover, Modeling, and Map Validation
3. Results
3.1. Image Masking and Validation
3.2. Time-Series Modeling and Metrics
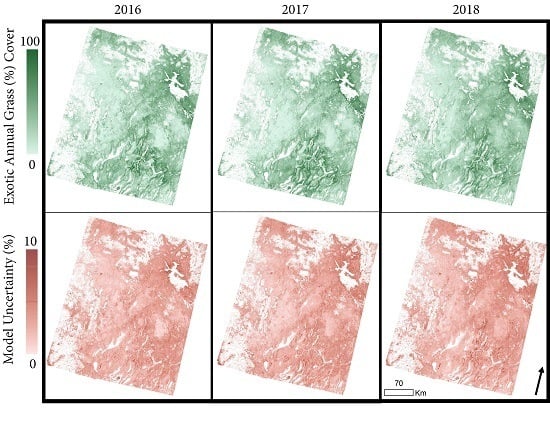
3.3. Exotic Annual Grass Cover Modeling, Mapping, and Validation
4. Discussion
4.1. Time Series Modeling, Metrics, and Image Masking
4.2. Exotic Annual Grass Cover Modeling, Mapping, and Validation
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Fusco, E.J.; Finn, J.T.; Balch, J.K.; Nagy, R.C.; Bradley, B.A. Invasive grasses increase fire occurrence and frequency across US ecoregions. Proc. Natl. Acad. Sci. USA 2019, 116, 23594–23599. [Google Scholar] [CrossRef]
- Balch, J.K.; Bradley, B.A.; D’Antonio, C.M.; Gomez-Dans, J. Introduced annual grass increases regional fire activity across the arid western USA (1980-2009). Glob. Chang. Biol. 2013, 19, 173–183. [Google Scholar] [CrossRef] [PubMed]
- Bradley, B.A.; Houghton, R.A.; Mustard, J.F.; Hamburg, S.P. Invasive grass reduces aboveground carbon stocks in shrublands of the Western US. Glob. Chang. Biol. 2006, 12, 1815–1822. [Google Scholar] [CrossRef] [Green Version]
- Chambers, J.C. Collaborative management and research in the Great Basin—Examining the issues and developing a framework for action. In Invasive Plant Species and the Great Basin; U.S. Department of Agriculture, Forest Service, Rocky Mountain Research Station: Fort Collins, CO, USA, 2008; pp. 38–41. [Google Scholar]
- Mack, R.N.; Pyke, D.A. The demography of Bromus Tectorum: Variation in time and space. J. Ecol. 1983, 71, 69–93. [Google Scholar] [CrossRef]
- Compagnoni, A.; Adler, P.B. Warming, competition, and Bromus tectorum population growth across an elevation gradient. Ecosphere 2014, 5, art121. [Google Scholar] [CrossRef]
- Chambers, J.C.; Roundy, B.A.; Blank, R.R.; Meyer, S.E.; Whittaker, A. What makes Great Basin sagebrush ecosystems invasible by Bromus tectorum? Ecol. Monogr. 2007, 77, 117–145. [Google Scholar] [CrossRef] [Green Version]
- Boyte, S.P.; Wylie, B.K.; Major, D.J. Cheatgrass percent cover change: Comparing recent estimates to climate change−driven predictions in the northern Great Basin. Rangel. Ecol. Manag. 2016, 69, 265–279. [Google Scholar] [CrossRef]
- Bradley, B.A.; Curtis, C.A.; Chambers, J.C. Bromus Response to Climate and Projected Changes with Climate Change. In Exotic Brome-Grasses in Arid and Semiarid Ecosystems of the Western US; Germino, M.J., Chambers, J.C., Brown, C.S., Eds.; Springer International Publishing: Cham, Switzerland, 2016; pp. 257–274. ISBN 978-3-319-24928-5. [Google Scholar]
- Abatzoglou, J.T.; Kolden, C.A. Climate Change in Western US Deserts: Potential for Increased Wildfire and Invasive Annual Grasses. Rangel. Ecol. Manag. 2011, 64, 471–478. [Google Scholar] [CrossRef]
- Chambers, J.C.; Bradley, B.A.; Brown, C.S.; D’Antonio, C.; Germino, M.J.; Grace, J.B.; Hardegree, S.P.; Miller, R.F.; Pyke, D.A. Resilience to Stress and Disturbance, and Resistance to Bromus tectorum L. Invasion in Cold Desert Shrublands of Western North America. Ecosystems 2014, 17, 360–375. [Google Scholar] [CrossRef]
- Bradley, B.A.; Mustard, J.F. Characterizing the landscape dynamics of an invasive plant and risk of invasion using remote sensing. Ecol. Appl. 2006, 16, 1132–1147. [Google Scholar] [CrossRef] [Green Version]
- Hobbs, R.J.; Humphries, S.E. An integrated approach to the ecology and management of plant invasions. Conserv. Biol. 1995, 9, 761–770. [Google Scholar] [CrossRef] [Green Version]
- Henderson, E.B.; Bell, D.M.; Gregory, M.J. Vegetation mapping to support greater sage-grouse habitat monitoring and management: Multi- or univariate approach? Ecosphere 2019, 10, e02838. [Google Scholar] [CrossRef]
- Jones, M.O.; Allred, B.W.; Naugle, D.E.; Maestas, J.D.; Donnelly, P.; Metz, L.J.; Karl, J.; Smith, R.; Bestelmeyer, B.; Boyd, C.; et al. Innovation in rangeland monitoring: Annual, 30 m, plant functional type percent cover maps for U.S. rangelands, 1984-2017. Ecosphere 2018, 9, e02430. [Google Scholar] [CrossRef]
- Xian, G.; Homer, C.; Rigge, M.; Shi, H.; Meyer, D. Characterization of shrubland ecosystem components as continuous fields in the northwest United States. Remote Sens. Environ. 2015, 168, 286–300. [Google Scholar] [CrossRef] [Green Version]
- Bradley, B.A. Remote detection of invasive plants: A review of spectral, textural and phenological approaches. Biol. Invasions 2013, 16, 1411–1425. [Google Scholar] [CrossRef]
- Boyte, S.P.; Wylie, B.K. Near-real-time cheatgrass percent cover in the northern Great Basin, USA, 2015. Rangelands 2016, 38, 278–284. [Google Scholar] [CrossRef] [Green Version]
- Boyte, S.P.; Wylie, B.K.; Major, D.J. Mapping and monitoring cheatgrass dieoff in rangelands of the northern Great Basin, USA. Rangel. Ecol. Manag. 2015, 68, 18–28. [Google Scholar] [CrossRef]
- Weisberg, P.J.; Dilts, T.E.; Baughman, O.W.; Meyer, S.E.; Leger, E.A.; Van Gunst, K.J.; Cleeves, L. Development of remote sensing indicators for mapping episodic die-off of an invasive annual grass (Bromus tectorum) from the Landsat archive. Ecol. Indic. 2017, 79, 173–181. [Google Scholar] [CrossRef]
- West, A.M.; Evangelista, P.H.; Jarnevich, C.S.; Kumar, S.; Swallow, A.; Luizza, M.W.; Chignell, S.M. Using multi-date satellite imagery to monitor invasive grass species distribution in post-wildfire landscapes: An iterative, adaptable approach that employs open-source data and software. Int. J. Appl. Earth Obs. Geoinf. 2017, 59, 135–146. [Google Scholar] [CrossRef]
- Ibáñez, I.; Silander, J.A., Jr.; Allen, J.M.; Treanor, S.A.; Wilson, A. Identifying hotspots for plant invasions and forecasting focal points of further spread. J. Appl. Ecol. 2009, 46, 1219–1228. [Google Scholar] [CrossRef] [Green Version]
- Vilà, M.; Ibáñez, I. Plant invasions in the landscape. Landsc. Ecol. 2011, 26, 461–472. [Google Scholar] [CrossRef]
- Clinton, N.E.; Potter, C.; Crabtree, B.; Genovese, V.; Gross, P.; Gong, P. Remote sensing-based time-series analysis of cheatgrass (Bromus tectorum L.) phenology. J. Environ. Qual. 2010, 39, 955–963. [Google Scholar] [CrossRef] [PubMed]
- Singh, N.; Glenn, N.F. Multitemporal spectral analysis for cheatgrass (Bromus tectorum) classification. Int. J. Remote Sens. 2009, 30, 3441–3462. [Google Scholar] [CrossRef]
- Bradley, B.A.; Mustard, J.F. Identifying land cover variability distinct from land cover change: Cheatgrass in the Great Basin. Remote Sens. Environ. 2005, 94, 204–213. [Google Scholar] [CrossRef]
- Claverie, M.; Ju, J.; Masek, J.G.; Dungan, J.L.; Vermote, E.F.; Roger, J.-C.; Skakun, S.V.; Justice, C. The Harmonized Landsat and Sentinel-2 surface reflectance data set. Remote Sens. Environ. 2018, 219, 145–161. [Google Scholar] [CrossRef]
- Omernik, J.M.; Griffith, G.E. Ecoregions of the conterminous United States: Evolution of a hierarchical spatial framework. Environ. Manag. 2014, 54, 1249–1266. [Google Scholar] [CrossRef]
- Trabucco, A.; Zomer, R.J. Global Aridity Index (Global-Aridity) and Global Potential Evapo-Transpiration (Global-PET) Geospatial Database 2019. Available online: https://cgiarcsi.community/2019/01/24/global-aridity-index-and-potential-evapotranspiration-climate-database-v2/ (accessed on 1 December 2019).
- Quinlan, J.R. C4.5: Programs for Machine Learning; Morgan Kaufmann Publishers, Inc.: San Francisco, CA, USA, 1993; ISBN 978-1-55860-238-0. [Google Scholar]
- Congalton, R.G. A review of assessing the accuracy of classifications of remotely sensed data. Remote Sens. Environ. 1991, 37, 35–46. [Google Scholar] [CrossRef]
- Pastick, N.; Wylie, B.; Wu, Z. Spatiotemporal analysis of Landsat-8 and Sentinel-2 data to support monitoring of dryland ecosystems. Remote Sens. 2018, 10, 791. [Google Scholar] [CrossRef] [Green Version]
- Hughes, M.; Hayes, D. Automated detection of cloud and cloud shadow in single-date Landsat imagery using neural networks and spatial post-processing. Remote Sens. 2014, 6, 4907–4926. [Google Scholar] [CrossRef] [Green Version]
- Jenkerson, C.B.; Maiersperger, T.; Schmidt, G. eMODIS: A User-Friendly Data Source; Open-File Report; U.S. Geological Survey: Reston, VA, USA, 2010.
- Araya, S.; Ostendorf, B.; Lyle, G.; Lewis, M. CropPhenology: An R package for extracting crop phenology from time series remotely sensed vegetation index imagery. Ecol. Inform. 2018, 46, 45–56. [Google Scholar] [CrossRef]
- Soil Survey Staff. Gridded Soil Survey Geographic (gSSURGO) Database for the Conterminous United States 2019. Available online: https://nrcs.app.box.com/v/soils/folder/94124173798 (accessed on 1 December 2019).
- Gesch, D.B.; Evans, G.A.; Oimoen, M.J.; Arundel, S. The National Elevation Dataset; American Society for Photogrammetry and Remote Sensing: Bethesda, MD, USA, 2018; pp. 83–110. [Google Scholar]
- Hull, A.C.; Pechanec, J.F. Cheatgrass: A challenge to range research. J. For. 1947, 45, 555–564. [Google Scholar]
- Stewart, G.; Hull, A.C. Cheatgrass (Bromus Tectorum L.)—An ecologic intruder in southern Idaho. Ecology 1949, 30, 58–74. [Google Scholar] [CrossRef]
- Rigge, M.; Shi, H.; Homer, C.; Danielson, P.; Granneman, B. Long-term trajectories of fractional component change in the Northern Great Basin, USA. Ecosphere 2019, 10, e02762. [Google Scholar] [CrossRef]
- White, J.C.; Wulder, M.A.; Hobart, G.W.; Luther, J.E.; Hermosilla, T.; Griffiths, P.; Coops, N.C.; Hall, R.J.; Hostert, P.; Dyk, A.; et al. Pixel-based image compositing for large-area dense time series applications and science. Can. J. Remote Sens. 2014, 40, 192–212. [Google Scholar] [CrossRef] [Green Version]
- Gamon, J.A.; Huemmrich, K.F.; Wong, C.Y.S.; Ensminger, I.; Garrity, S.; Hollinger, D.Y.; Noormets, A.; Peñuelas, J. A remotely sensed pigment index reveals photosynthetic phenology in evergreen conifers. Proc. Natl. Acad. Sci. USA 2016, 113, 13087. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Robinson, P.N.; Allred, W.B.; Jones, O.M.; Moreno, A.; Kimball, S.J.; Naugle, E.D.; Erickson, A.T.; Richardson, D.A. A Dynamic Landsat Derived Normalized Difference Vegetation Index (NDVI) Product for the Conterminous United States. Remote Sens. 2017, 9, 863. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Q.; Rover, J.; Brown, J.; Worstell, B.; Howard, D.; Wu, Z.; Gallant, L.A.; Rundquist, B.; Burke, M. Monitoring Landscape Dynamics in Central U.S. Grasslands with Harmonized Landsat-8 and Sentinel-2 Time Series Data. Remote Sens. 2019, 11, 328. [Google Scholar] [CrossRef] [Green Version]
- Yang, L.; Jin, S.; Danielson, P.; Homer, C.; Gass, L.; Bender, S.M.; Case, A.; Costello, C.; Dewitz, J.; Fry, J.; et al. A new generation of the United States National Land Cover Database: Requirements, research priorities, design, and implementation strategies. ISPRS J. Photogramm. Remote Sens. 2018, 146, 108–123. [Google Scholar] [CrossRef]
- Homer, C.G.; Aldridge, C.L.; Meyer, D.K.; Schell, S.J. Multi-scale remote sensing sagebrush characterization with regression trees over Wyoming, USA: Laying a foundation for monitoring. Int. J. Appl. Earth Obs. Geoinf. 2012, 14, 233–244. [Google Scholar] [CrossRef]
- Smith, W.K.; Dannenberg, M.P.; Yan, D.; Herrmann, S.; Barnes, M.L.; Barron-Gafford, G.A.; Biederman, J.A.; Ferrenberg, S.; Fox, A.M.; Hudson, A.; et al. Remote sensing of dryland ecosystem structure and function: Progress, challenges, and opportunities. Remote Sens. Environ. 2019, 233, 111401. [Google Scholar] [CrossRef]
- Qiu, S.; Zhu, Z.; He, B. Fmask 4.0: Improved cloud and cloud shadow detection in Landsats 4–8 and Sentinel-2 imagery. Remote Sens. Environ. 2019, 231, 111205. [Google Scholar] [CrossRef]
- Ganguly, S.; Friedl, M.A.; Tan, B.; Zhang, X.; Verma, M. Land surface phenology from MODIS: Characterization of the Collection 5 global land cover dynamics product. Remote Sens. Environ. 2010, 114, 1805–1816. [Google Scholar] [CrossRef] [Green Version]
- White, M.A.; De Beurs, K.M.; Didan, K.; Inouye, D.W.; Richardson, A.D.; Jensen, O.P.; O’Keefe, J.; Zhang, G.; Nemani, R.R.; Van Leeuwn, W.J.D.; et al. Intercomparison, interpretation, and assessment of spring phenology in North America estimated from remote sensing for 1982–2006. Glob. Chang. Biol. 2009, 15, 2335–2359. [Google Scholar] [CrossRef]
- Kokaly, R.F. Detecting Cheatgrass on the Colorado Plateau Using Landsat Data: A Tutorial for the DESI Software; Open-File Report; U.S. Geological Survey: Reston, VA, USA, 2011.
- Hansen, M.C. Classification Trees and Mixed Pixel Training Data. In Remote Sensing of Land Cover: Principles and Applications; Chandra, G., Ed.; Taylor and Francis: New York, NY, USA, 2012. [Google Scholar]
- Dahal, D.; Wylie, B.K.; Parajuli, S.; Pastick, N.J. Fractional Estimates of Invasive Annual Grass cover in Dryland Ecosystems of Western United States (2016–2018); U.S. Geological Survey: Reston, VA, USA, 2020.
- Shinneman, D.J.; Germino, M.J.; Pilliod, D.S.; Aldridge, C.L.; Vaillant, N.M.; Coates, P.S. The ecological uncertainty of wildfire fuel breaks: Examples from the sagebrush steppe. Front. Ecol. Environ. 2019, 17, 279–288. [Google Scholar] [CrossRef]







| Standard HLS Mask | Decision Tree Mask | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Landsat-8 | Reference | Landsat-8 | Reference | ||||||||
| Clear | Non-Clear | Total | User’s Accuracy | Clear | Non-Clear | Total | User’s Accuracy | ||||
| Map | Clear | 577 | 73 | 650 | 89 ± 2% | Map | Clear | 686 | 131 | 817 | 84 ± 3% |
| Non-Clear | 168 | 482 | 650 | 74 ± 3% | Non-Clear | 59 | 424 | 483 | 88 ± 3% | ||
| Total | 745 | 555 | Total | 745 | 555 | ||||||
| Producer’s Accuracy | 84 ± 2% | 81 ± 3% | Producer’s Accuracy | 92 ± 2% | 76 ± 3% | ||||||
| Overall Accuracy | 83 ± 2% | Overall Accuracy | 85 ± 2% | ||||||||
| Sentinel-2 | Reference | Sentinel-2 | Reference | ||||||||
| Clear | Non-Clear | Total | User’s Accuracy | Clear | Non-Clear | Total | User’s Accuracy | ||||
| Map | Clear | 550 | 100 | 650 | 85 ± 3% | Map | Clear | 616 | 116 | 732 | 84 ± 3% |
| Non-Clear | 195 | 455 | 650 | 70 ± 4% | Non-Clear | 129 | 439 | 568 | 77 ± 4% | ||
| Total | 745 | 555 | Total | 745 | 555 | ||||||
| Producer’s Accuracy | 78 ± 2% | 78 ± 3% | Producer’s Accuracy | 83 ± 2% | 79 ± 3% | ||||||
| Overall Accuracy | 78 ± 2% | Overall Accuracy | 81 ± 2% | ||||||||
| Ecoregion | % Study Area | Year | eMODIS and HLSRT (r l) | eMODIS and HLSint (r l) |
|---|---|---|---|---|
| Blue Mountains | 17 | 2016 | 0.86 2 | 0.86 2 |
| 2017 | 0.77 2 | 0.792 | ||
| 2018 | 0.934 | 0.89 3 | ||
| Cascades | 4 | 2016 | 0.37 1 | 0.590 |
| 2017 | 0.59 2 | 0.750 | ||
| 2018 | 0.76 2 | 0.771 | ||
| Central Basin and Range | 22 | 2016 | 0.91 1 | 0.921 |
| 2017 | 0.91 2 | 0.91 1 | ||
| 2018 | 0.923 | 0.90 2 | ||
| Central California Foothills and Coastal Mountains | 0.3 | 2016 | 0.94 1 | 0.961 |
| 2017 | 0.96 1 | 0.970 | ||
| 2018 | 0.95 1 | 0.980 | ||
| Columbia Plateau | 0.6 | 2016 | 0.981 | 0.96 1 |
| 2017 | 0.971 | 0.94 1 | ||
| 2018 | 0.922 | 0.85 2 | ||
| Eastern Cascades and Foothills | 11.9 | 2016 | 0.822 | 0.80 2 |
| 2017 | 0.554 | 0.54 5 | ||
| 2018 | 0.832 | 0.75 2 | ||
| Idaho Batholith | 3.3 | 2016 | 0.881 | 0.86 2 |
| 2017 | 0.752 | 0.74 2 | ||
| 2018 | 0.874 | 0.83 3 | ||
| Northern Basin and Range | 33.6 | 2016 | 0.90 1 | 0.911 |
| 2017 | 0.74 2 | 0.762 | ||
| 2018 | 0.923 | 0.90 2 | ||
| Sierra Nevada | 2.5 | 2016 | 0.63 1 | 0.781 |
| 2017 | 0.50 4 | 0.692 | ||
| 2018 | 0.76 3 | 0.793 | ||
| Snake River Plain | 5.2 | 2016 | 0.98 1 | 0.98 1 |
| 2017 | 0.861 | 0.83 2 | ||
| 2018 | 0.95 3 | 0.982 | ||
| Overall | 100 | - | 0.911 | 0.87 2 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pastick, N.J.; Dahal, D.; Wylie, B.K.; Parajuli, S.; Boyte, S.P.; Wu, Z. Characterizing Land Surface Phenology and Exotic Annual Grasses in Dryland Ecosystems Using Landsat and Sentinel-2 Data in Harmony. Remote Sens. 2020, 12, 725. https://0-doi-org.brum.beds.ac.uk/10.3390/rs12040725
Pastick NJ, Dahal D, Wylie BK, Parajuli S, Boyte SP, Wu Z. Characterizing Land Surface Phenology and Exotic Annual Grasses in Dryland Ecosystems Using Landsat and Sentinel-2 Data in Harmony. Remote Sensing. 2020; 12(4):725. https://0-doi-org.brum.beds.ac.uk/10.3390/rs12040725
Chicago/Turabian StylePastick, Neal J., Devendra Dahal, Bruce K. Wylie, Sujan Parajuli, Stephen P. Boyte, and Zhouting Wu. 2020. "Characterizing Land Surface Phenology and Exotic Annual Grasses in Dryland Ecosystems Using Landsat and Sentinel-2 Data in Harmony" Remote Sensing 12, no. 4: 725. https://0-doi-org.brum.beds.ac.uk/10.3390/rs12040725