Mapping Floristic Patterns of Trees in Peruvian Amazonia Using Remote Sensing and Machine Learning
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Area
2.2. Floristic Data
2.3. Landsat and Environmental Data
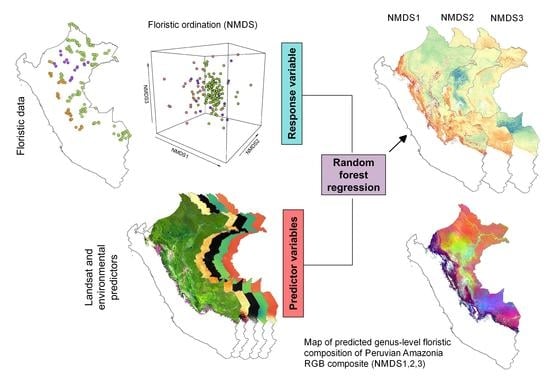
2.4. Data Analyses
3. Results
3.1. Floristic Patterns and Their Correlation with Landsat and Environmental Variables
3.2. Predicting Tree Community Composition at the Genus Level Throughout Peruvian Amazonia
3.3. Indicator Analysis
4. Discussion
4.1. Correlates of Floristic Patterns
4.2. Predicting Floristic Composition of Trees Using Landsat Reflectance Values
4.3. Interpretation of the Floristic Map and Indicator Genera
4.4. Practical Implications and Recommendations
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Data Availability
References
- Coe, M.T.; Costa, M.H.; Soares-Filho, B.S. The influence of historical and potential future deforestation on the stream flow of the Amazon River—Land surface processes and atmospheric feedbacks. J. Hydrol. 2009, 369, 165–174. [Google Scholar] [CrossRef]
- Hopkins, M.J.G. Modelling the known and unknown plant biodiversity of the Amazon Basin. J. Biogeogr. 2007, 34, 1400–1411. [Google Scholar] [CrossRef]
- Schulman, L.; Toivonen, T.; Ruokolainen, K. Analysing botanical collecting effort in Amazonia and correcting for it in species range estimation. J. Biogeogr. 2007, 34, 1388–1399. [Google Scholar] [CrossRef]
- Sousa-Baena, M.S.; Garcia, L.C.; Peterson, A.T. Completeness of digital accessible knowledge of the plants of Brazil and priorities for survey and inventory. Divers. Distrib. 2014, 20, 369–381. [Google Scholar] [CrossRef]
- He, K.S.; Bradley, B.A.; Cord, A.F.; Rocchini, D.; Tuanmu, M.-N.; Schmidtlein, S.; Turner, W.; Wegmann, M.; Pettorelli, N. Will remote sensing shape the next generation of species distribution models? Remote Sens. Ecol. Conserv. 2015, 1, 4–18. [Google Scholar] [CrossRef] [Green Version]
- Turner, W. Sensing biodiversity. Science 2014, 346, 301–302. [Google Scholar] [CrossRef]
- Rocchini, D.; Luque, S.; Pettorelli, N.; Bastin, L.; Doktor, D.; Faedi, N.; Feilhauer, H.; Féret, J.-B.; Foody, G.M.; Gavish, Y.; et al. Measuring β-diversity by remote sensing: A challenge for biodiversity monitoring. Methods Ecol. Evol. 2018, 9, 1787–1798. [Google Scholar] [CrossRef] [Green Version]
- Rocchini, D.; Boyd, D.S.; Féret, J.-B.; Foody, G.M.; He, K.S.; Lausch, A.; Nagendra, H.; Wegmann, M.; Pettorelli, N. Satellite remote sensing to monitor species diversity: Potential and pitfalls. Remote Sens. Ecol. Conserv. 2016, 2, 25–36. [Google Scholar] [CrossRef]
- Asner, G.P.; Anderson, C.B.; Martin, R.E.; Knapp, D.E.; Tupayachi, R.; Sinca, F.; Malhi, Y. Landscape-scale changes in forest structure and functional traits along an Andes-to-Amazon elevation gradient. Biogeosciences 2014, 11, 843–856. [Google Scholar] [CrossRef] [Green Version]
- Asner, G.P.; Martin, R.E.; Knapp, D.E.; Tupayachi, R.; Anderson, C.B.; Sinca, F.; Vaughn, N.R.; Llactayo, W. Airborne laser-guided imaging spectroscopy to map forest trait diversity and guide conservation. Science 2017, 355, 385–389. [Google Scholar] [CrossRef]
- Asner, G.P.; Martin, R.E.; Tupayachi, R.; Llactayo, W. Conservation assessment of the Peruvian Andes and Amazon based on mapped forest functional diversity. Biol. Conserv. 2017, 210, 80–88. [Google Scholar] [CrossRef]
- Asner, G.P.; Martin, R.E. Airborne spectranomics: Mapping canopy chemical and taxonomic diversity in tropical forests. Front. Ecol. Environ. 2009, 7, 269–276. [Google Scholar] [CrossRef] [Green Version]
- Tuomisto, H.; Van doninck, J.; Ruokolainen, K.; Moulatlet, G.M.; Figueiredo, F.O.G.; Sirén, A.; Cárdenas, G.; Lehtonen, S.; Zuquim, G. Discovering floristic and geoecological gradients across Amazonia. J. Biogeogr. 2019, 46, 1734–1748. [Google Scholar] [CrossRef]
- Saatchi, S.; Buermann, W.; ter Steege, H.; Mori, S.; Smith, T.B. Modeling distribution of Amazonian tree species and diversity using remote sensing measurements. Remote Sens. Environ. 2008, 112, 2000–2017. [Google Scholar] [CrossRef]
- ter Steege, H.; Sabatier, D.; Castellanos, H.; Andel, T.V.; Duivenvoorden, J.; Oliveira, A.A.D.; Ek, R.; Lilwah, R.; Maas, P.; Mori, S. An analysis of the floristic composition and diversity of Amazonian forests including those of the Guiana Shield. J. Trop. Ecol. 2000, 16, 801–828. [Google Scholar] [CrossRef] [Green Version]
- ter Steege, H.; Pitman, N.; Sabatier, D.; Castellanos, H.; van der Hout, P.; Daly, D.C.; Silveira, M.; Phillips, O.; Vasquez, R.; van Andel, T.; et al. A spatial model of tree α-diversity and tree density for the Amazon. Biodivers. Conserv. 2003, 12, 2255–2277. [Google Scholar] [CrossRef]
- ter Steege, H.; Pitman, N.C.A.; Phillips, O.L.; Chave, J.; Sabatier, D.; Duque, A.; Molino, J.-F.; Prévost, M.-F.; Spichiger, R.; Castellanos, H.; et al. Continental-scale patterns of canopy tree composition and function across Amazonia. Nature 2006, 443, 444–447. [Google Scholar] [CrossRef] [PubMed]
- Chaves, P.P.; Ruokolainen, K.; Tuomisto, H. Using remote sensing to model tree species distribution in Peruvian lowland Amazonia. Biotropica 2018, 50, 758–767. [Google Scholar] [CrossRef]
- Tuomisto, H.; Ruokolainen, K.; Kalliola, R.; Linna, A.; Danjoy, W.; Rodriguez, Z. Dissecting Amazonian Biodiversity. Science 1995, 269, 63–66. [Google Scholar] [CrossRef]
- Asner, G.P.; Knapp, D.E.; Martin, R.E.; Tupayachi, R.; Anderson, C.B.; Mascaro, J.; Sinca, F.; Chadwick, K.D.; Sousan, S.; Higgins, M.; et al. The High-Resolution Carbon Geography of Perú: A Collaborative Report of the Carnegie Airborne Observatory and the Ministry of Environment of Perú; Carnegie Institution for Science: Washington, DC, USA, 2014. [Google Scholar]
- Saatchi, S.; Malhi, Y.; Zutta, B.; Buermann, W.; Anderson, L.O.; Araujo, A.M.; Phillips, O.L.; Peacock, J.; ter Steege, H.; Lopez Gonzalez, G.; et al. Mapping landscape scale variations of forest structure, biomass, and productivity in Amazonia. Biogeosciences Discuss. 2009, 6, 5461–5505. [Google Scholar] [CrossRef] [Green Version]
- Rödig, E.; Knapp, N.; Fischer, R.; Bohn, F.J.; Dubayah, R.; Tang, H.; Huth, A. From small-scale forest structure to Amazon-wide carbon estimates. Nat. Commun. 2019, 10, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Zuquim, G.; Tuomisto, H.; Jones, M.M.; Prado, J.; Figueiredo, F.O.G.; Moulatlet, G.M.; Costa, F.R.C.; Quesada, C.A.; Emilio, T. Predicting environmental gradients with fern species composition in Brazilian Amazonia. J. Veg. Sci. 2014, 25, 1195–1207. [Google Scholar] [CrossRef]
- Higgins, M.A.; Ruokolainen, K.; Tuomisto, H.; Llerena, N.; Cardenas, G.; Phillips, O.L.; Vásquez, R.; Räsänen, M. Geological control of floristic composition in Amazonian forests. J. Biogeogr. 2011, 38, 2136–2149. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tuomisto, H.; Poulsen, A.D.; Ruokolainen, K.; Moran, R.C.; Quintana, C.; Celi, J.; Cañas, G. Linking floristic patterns with soil heterogeneity and satellite imagery in Ecuadorian Amazonia. Ecol. Appl. 2003, 13, 352–371. [Google Scholar] [CrossRef]
- Tuomisto, H.; Ruokolainen, K.; Aguilar, M.; Sarmiento, A. Floristic patterns along a 43-km long transect in an Amazonian rain forest. J. Ecol. 2003, 91, 743–756. [Google Scholar] [CrossRef]
- Zuquim, G.; Tuomisto, H.; Costa, F.R.C.; Prado, J.; Magnusson, W.E.; Pimentel, T.; Braga-Neto, R.; Figueiredo, F.O.G. Broad Scale Distribution of Ferns and Lycophytes along Environmental Gradients in Central and Northern Amazonia, Brazil. Biotropica 2012, 44, 752–762. [Google Scholar] [CrossRef]
- Figueiredo, F.O.G.; Zuquim, G.; Tuomisto, H.; Moulatlet, G.M.; Balslev, H.; Costa, F.R.C. Beyond climate control on species range: The importance of soil data to predict distribution of Amazonian plant species. J. Biogeogr. 2018, 45, 190–200. [Google Scholar] [CrossRef]
- Vormisto, J.; Svenning, J.-C.; Hall, P.; Balslev, H. Diversity and dominance in palm (Arecaceae) communities in terra firme forests in the western Amazon basin. J. Ecol. 2004, 92, 577–588. [Google Scholar] [CrossRef]
- Kristiansen, T.; Svenning, J.-C.; Eiserhardt, W.L.; Pedersen, D.; Brix, H.; Munch Kristiansen, S.; Knadel, M.; Grández, C.; Balslev, H. Environment versus dispersal in the assembly of western Amazonian palm communities. J. Biogeogr. 2012, 39, 1318–1332. [Google Scholar] [CrossRef]
- Cámara-Leret, R.; Tuomisto, H.; Ruokolainen, K.; Balslev, H.; Munch Kristiansen, S. Modelling responses of western Amazonian palms to soil nutrients. J. Ecol. 2017, 105, 367–381. [Google Scholar] [CrossRef]
- Wittmann, F.; Schongart, J.; Montero, J.C.; Motzer, T.; Junk, W.J.; Piedade, M.T.F.; Queiroz, H.L.; Worbes, M. Tree species composition and diversity gradients in white-water forests across the Amazon Basin. J. Biogeogr. 2006, 33, 1334–1347. [Google Scholar] [CrossRef]
- Macía, M.J. Spatial distribution and floristic composition of trees and lianas in different forest types of an Amazonian rainforest. Plant Ecol. 2011, 212, 1159–1177. [Google Scholar] [CrossRef]
- Toledo, M.; Poorter, L.; Peña-Claros, M.; Alarcón, A.; Balcázar, J.; Chuviña, J.; Leaño, C.; Licona, J.C.; ter Steege, H.; Bongers, F. Patterns and Determinants of Floristic Variation across Lowland Forests of Bolivia. Biotropica 2011, 43, 405–413. [Google Scholar] [CrossRef]
- Toledo, M.; Peña-Claros, M.; Bongers, F.; Alarcón, A.; Balcázar, J.; Chuviña, J.; Leaño, C.; Licona, J.C.; Poorter, L. Distribution patterns of tropical woody species in response to climatic and edaphic gradients. J. Ecol. 2012, 100, 253–263. [Google Scholar] [CrossRef]
- Ruokolainen, K.; Tuomisto, H.; Macía, M.J.; Higgins, M.A.; Yli-Halla, M. Are floristic and edaphic patterns in Amazonian rain forests congruent for trees, pteridophytes and Melastomataceae? J. Trop. Ecol. 2007, 23, 13–25. [Google Scholar] [CrossRef] [Green Version]
- Baldeck, C.A.; Harms, K.E.; Yavitt, J.B.; John, R.; Turner, B.L.; Valencia, R.; Navarrete, H.; Davies, S.J.; Chuyong, G.B.; Kenfack, D.; et al. Soil resources and topography shape local tree community structure in tropical forests. Proc. R. Soc. Lond. B Biol. Sci. 2013, 280, 20122532. [Google Scholar] [CrossRef]
- Assis, R.L.; Haugaasen, T.; Schöngart, J.; Montero, J.C.; Piedade, M.T.F.; Wittmann, F. Patterns of tree diversity and composition in Amazonian floodplain paleo-várzea forest. J. Veg. Sci. 2015, 26, 312–322. [Google Scholar] [CrossRef]
- Baldeck, C.A.; Tupayachi, R.; Sinca, F.; Jaramillo, N.; Asner, G.P. Environmental drivers of tree community turnover in western Amazonian forests. Ecography 2016, 39, 1089–1099. [Google Scholar] [CrossRef]
- Phillips, O.L.; Vargas, P.N.; Monteagudo, A.L.; Cruz, A.P.; Zans, M.-E.C.; Sánchez, W.G.; Yli-Halla, M.; Rose, S. Habitat association among Amazonian tree species: A landscape-scale approach. J. Ecol. 2003, 91, 757–775. [Google Scholar] [CrossRef] [Green Version]
- Jones, M.M.; Ferrier, S.; Condit, R.; Manion, G.; Aguilar, S.; Pérez, R. Strong congruence in tree and fern community turnover in response to soils and climate in central Panama. J. Ecol. 2013, 101, 506–516. [Google Scholar] [CrossRef]
- Tuomisto, H.; Moulatlet, G.M.; Balslev, H.; Emilio, T.; Figueiredo, F.O.G.; Pedersen, D.; Ruokolainen, K. A compositional turnover zone of biogeographical magnitude within lowland Amazonia. J. Biogeogr. 2016, 43, 2400–2411. [Google Scholar] [CrossRef]
- Moulatlet, G.M.; Zuquim, G.; Figueiredo, F.O.G.; Lehtonen, S.; Emilio, T.; Ruokolainen, K.; Tuomisto, H. Using digital soil maps to infer edaphic affinities of plant species in Amazonia: Problems and prospects. Ecol. Evol. 2017, 7, 8463–8477. [Google Scholar] [CrossRef] [Green Version]
- Soria-Auza, R.W.; Kessler, M.; Bach, K.; Barajas-Barbosa, P.M.; Lehnert, M.; Herzog, S.K.; Böhner, J. Impact of the quality of climate models for modelling species occurrences in countries with poor climatic documentation: A case study from Bolivia. Ecol. Model. 2010, 221, 1221–1229. [Google Scholar] [CrossRef]
- Sirén, A.; Tuomisto, H.; Navarrete, H. Mapping environmental variation in lowland Amazonian rainforests using remote sensing and floristic data. Int. J. Remote Sens. 2013, 34, 1561–1575. [Google Scholar] [CrossRef]
- Muro, J.; Van doninck, J.; Tuomisto, H.; Higgins, M.A.; Moulatlet, G.M.; Ruokolainen, K. Floristic composition and across-track reflectance gradient in Landsat images over Amazonian forests. ISPRS J. Photogramm. Remote Sens. 2016, 119, 361–372. [Google Scholar] [CrossRef] [Green Version]
- Higgins, M.A.; Asner, G.P.; Perez, E.; Elespuru, N.; Tuomisto, H.; Ruokolainen, K.; Alonso, A. Use of Landsat and SRTM Data to Detect Broad-Scale Biodiversity Patterns in Northwestern Amazonia. Remote Sens. 2012, 4, 2401–2418. [Google Scholar] [CrossRef] [Green Version]
- Thessler, S.; Ruokolainen, K.; Tuomisto, H.; Tomppo, E. Mapping gradual landscape-scale floristic changes in Amazonian primary rain forests by combining ordination and remote sensing. Glob. Ecol. Biogeogr. 2005, 14, 315–325. [Google Scholar] [CrossRef]
- Van doninck, J.; Tuomisto, H. A Landsat composite covering all Amazonia for applications in ecology and conservation. Remote Sens. Ecol. Conserv. 2018, 4, 197–210. [Google Scholar] [CrossRef] [Green Version]
- Van doninck, J.; Jones, M.M.; Zuquim, G.; Ruokolainen, K.; Moulatlet, G.M.; Sirén, A.; Cárdenas, G.; Lehtonen, S.; Tuomisto, H. Multispectral canopy reflectance improves spatial distribution models of Amazonian understory species. Ecography 2020, 43, 128–137. [Google Scholar] [CrossRef]
- Honorio Coronado, E.N.; Baker, T.R.; Phillips, O.L.; Pitman, N.C.A.; Pennington, R.T.; Vásquez Martínez, R.; Monteagudo, A.; Mogollón, H.; Dávila Cardozo, N.; Ríos, M.; et al. Multi-scale comparisons of tree composition in Amazonian terra firme forests. Biogeosciences 2009, 6, 2719–2731. [Google Scholar] [CrossRef] [Green Version]
- Cayuela, L.; de la Cruz, M.; Ruokolainen, K. A method to incorporate the effect of taxonomic uncertainty on multivariate analyses of ecological data. Ecography 2011, 34, 94–102. [Google Scholar] [CrossRef] [Green Version]
- Higgins, M.A.; Ruokolainen, K. Rapid Tropical Forest Inventory: A Comparison of Techniques Based on Inventory Data from Western Amazonia. Conserv. Biol. 2004, 18, 799–811. [Google Scholar] [CrossRef]
- MINAM Mapa de Propuesta Técnica de Límites Amazónicos. Available online: http://geoservidor.minam.gob.pe/recursos/intercambio-de-datos/ (accessed on 19 April 2020).
- Karger, D.N.; Conrad, O.; Böhner, J.; Kawohl, T.; Kreft, H.; Soria-Auza, R.W.; Zimmermann, N.E.; Linder, H.P.; Kessler, M. Climatologies at high resolution for the earth’s land surface areas. Sci. Data 2017, 4, 170122. [Google Scholar] [CrossRef] [Green Version]
- SERFOR Memoria Descriptiva del Mapa de Ecozonas, Inventario Nacional Forestal y de Fauna Silvestre (INFFS)-Perú. Available online: https://sinia.minam.gob.pe/documentos/memoria-descriptiva-mapa-ecozonas-inventario-nacional-forestal-fauna (accessed on 19 March 2020).
- SERFOR Marco Metodológico del Inventario Nacional Forestal y de Fauna Silvestre—Perú. Available online: https://www.serfor.gob.pe/wp-content/uploads/2016/05/Marco-metodologico-INFFS.pdf. (accessed on 19 March 2020).
- Van doninck, J.; Tuomisto, H. BRDF correction for Landsat TM/ETM+ data over Amazonian forests. In Proceedings of the 2nd EARSeL International Workshop on Temporal Analysis of Satellite Images, Stockholm, Sweden, 17–19 June 2015; pp. 349–354. [Google Scholar]
- Van doninck, J.; Tuomisto, H. Evaluation of directional normalization methods for Landsat TM/ETM+ over primary Amazonian lowland forests. Int. J. Appl. Earth Obs. Geoinf. 2017, 58, 249–263. [Google Scholar] [CrossRef]
- Van doninck, J.; Tuomisto, H. Influence of Compositing Criterion and Data Availability on Pixel-Based Landsat TM/ETM+ Image Compositing over Amazonian Forests. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2017, 10, 857–867. [Google Scholar] [CrossRef]
- Hengl, T.; de Jesus, J.M.; Heuvelink, G.B.M.; Gonzalez, M.R.; Kilibarda, M.; Blagotić, A.; Shangguan, W.; Wright, M.N.; Geng, X.; Bauer-Marschallinger, B.; et al. SoilGrids250m: Global gridded soil information based on machine learning. PLoS ONE 2017, 12, e0169748. [Google Scholar] [CrossRef] [Green Version]
- De’ath, G. Extended dissimilarity: A method of robust estimation of ecological distances from high beta diversity data. Plant Ecol. 1999, 144, 191–199. [Google Scholar] [CrossRef]
- Tuomisto, H.; Ruokolainen, L.; Ruokolainen, K. Modelling niche and neutral dynamics: On the ecological interpretation of variation partitioning results. Ecography 2012, 35, 961–971. [Google Scholar] [CrossRef]
- Legendre, P.; Legendre, L. Numerical Ecology, 3rd ed.; Developments in Environmental Modelling; Elsevier: Amsterdam, The Netherlands; Oxford, UK, 2012; Volume 24, ISBN 978-0-444-53868-0. [Google Scholar]
- Kuhn, M.; Johnson, K. Applied Predictive Modeling; Springer: New York, NY, USA, 2013; ISBN 978-1-4614-6848-6. [Google Scholar]
- Meyer, H.; Reudenbach, C.; Hengl, T.; Katurji, M.; Nauss, T. Improving performance of spatio-temporal machine learning models using forward feature selection and target-oriented validation. Environ. Model. Softw. 2018, 101, 1–9. [Google Scholar] [CrossRef]
- Meyer, H.; Reudenbach, C.; Wöllauer, S.; Nauss, T. Importance of spatial predictor variable selection in machine learning applications—Moving from data reproduction to spatial prediction. Ecol. Model. 2019, 411, 108815. [Google Scholar] [CrossRef] [Green Version]
- Roberts, D.R.; Bahn, V.; Ciuti, S.; Boyce, M.S.; Elith, J.; Guillera-Arroita, G.; Hauenstein, S.; Lahoz-Monfort, J.J.; Schröder, B.; Thuiller, W.; et al. Cross-validation strategies for data with temporal, spatial, hierarchical, or phylogenetic structure. Ecography 2017, 40, 913–929. [Google Scholar] [CrossRef]
- Schratz, P.; Muenchow, J.; Iturritxa, E.; Richter, J.; Brenning, A. Hyperparameter tuning and performance assessment of statistical and machine-learning algorithms using spatial data. Ecol. Model. 2019, 406, 109–120. [Google Scholar] [CrossRef] [Green Version]
- Karasiak, N.; Dejoux, J.-F.; Fauvel, M.; Willm, J.; Monteil, C.; Sheeren, D. Statistical Stability and Spatial Instability in Mapping Forest Tree Species by Comparing 9 Years of Satellite Image Time Series. Remote Sens. 2019, 11, 2512. [Google Scholar] [CrossRef] [Green Version]
- De Cáceres, M.; Legendre, P.; Valencia, R.; Cao, M.; Chang, L.-W.; Chuyong, G.; Condit, R.; Hao, Z.; Hsieh, C.-F.; Hubbell, S.; et al. The variation of tree beta diversity across a global network of forest plots: Beta diversity across a network of forest plots. Glob. Ecol. Biogeogr. 2012, 21, 1191–1202. [Google Scholar] [CrossRef]
- Dufrêne, M.; Legendre, P. Species assemblages and indicator species: The need for a flexible asymmetrical approach. Ecol. Monogr. 1997, 67, 345–366. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2018. [Google Scholar]
- Oksanen, J.; Blanchet, F.G.; Friendly, M.; Kindt, R.; Legendre, P.; McGlinn, D.; Minchin, P.R.; O’Hara, R.B.; Simpson, G.L.; Solymos, P.; et al. vegan: Community Ecology Package; R package version, 2018; Available online: https://cran.r-project.org/web/packages/vegan/index.html (accessed on 10 May 2020).
- De Cáceres, M.; Jansen, F. indicspecies: Relationship between Species and Groups of Sites. R Package; R Package Version, 2015; Available online: https://cran.r-project.org/web/packages/indicspecies/index.html (accessed on 10 May 2020).
- Hijmans, R.J. raster: Geographic Data Analysis and Modeling; R Package Version, 2017; Available online: https://cran.r-project.org/web/packages/raster/index.html (accessed on 10 May 2020).
- Wing, M.K.C.J.; Weston, S.; Williams, A.; Keefer, C.; Engelhardt, A.; Cooper, T.; Mayer, Z.; Kenkel, B.; R Core Team; Benesty, M.; et al. caret: Classification and Regression Training; R Package Version, 2019; Available online: https://cran.r-project.org/web/packages/caret/index.html (accessed on 10 May 2020).
- Meyer, H.; Reudenbach, C.; Ludwig, M.; Nauss, T. CAST: “caret” Applications for Spatial-Temporal Models; R Package Version, 2018; Available online: http://search.r-project.org/library/CAST/html/CAST.html (accessed on 10 May 2020).
- Maechler, M.; Rousseeuw, P.; Struyf, A.; Hubert, M.; Hornik, K. cluster: Cluster Analysis Basics and Extensions; R Package Version, 2016; Available online: https://cran.r-project.org/web/packages/cluster/index.html (accessed on 10 May 2020).
- Lähteenoja, O.; Page, S. High diversity of tropical peatland ecosystem types in the Pastaza-Marañón basin, Peruvian Amazonia. J. Geophys. Res. 2011, 116, G02025. [Google Scholar] [CrossRef] [Green Version]
- de Carvalho, A.L.; Nelson, B.W.; Bianchini, M.C.; Plagnol, D.; Kuplich, T.M.; Daly, D.C. Bamboo-Dominated Forests of the Southwest Amazon: Detection, Spatial Extent, Life Cycle Length and Flowering Waves. PLoS ONE 2013, 8, e54852. [Google Scholar] [CrossRef]
- MINAM Mapa Nacional de Ecosistemas del Perú. Available online: https://sinia.minam.gob.pe/mapas/mapa-nacional-ecosistemas-peru (accessed on 19 March 2020).
- Chust, G.; Chave, J.; Condit, R.; Aguilar, S.; Lao, S.; Pérez, R. Determinants and spatial modeling of tree β-diversity in a tropical forest landscape in Panama. J. Veg. Sci. 2006, 17, 83–92. [Google Scholar] [CrossRef]
- Condit, R.; Engelbrecht, B.M.J.; Pino, D.; Pérez, R.; Turner, B.L. Species distributions in response to individual soil nutrients and seasonal drought across a community of tropical trees. Proc. Natl. Acad. Sci. USA 2013, 110, 5064–5068. [Google Scholar] [CrossRef] [Green Version]
- Asner, G.P.; Martin, R.E.; Anderson, C.B.; Knapp, D.E. Quantifying forest canopy traits: Imaging spectroscopy versus field survey. Remote Sens. Environ. 2015, 158, 15–27. [Google Scholar] [CrossRef]
- Asner, G.P.; Anderson, C.B.; Martin, R.E.; Tupayachi, R.; Knapp, D.E.; Sinca, F. Landscape biogeochemistry reflected in shifting distributions of chemical traits in the Amazon forest canopy. Nat. Geosci. 2015, 8, 567–573. [Google Scholar] [CrossRef]
- Baldeck, C.A.; Asner, G.P. Estimating Vegetation Beta Diversity from Airborne Imaging Spectroscopy and Unsupervised Clustering. Remote Sens. 2013, 5, 2057–2071. [Google Scholar] [CrossRef] [Green Version]
- Baldeck, C.A.; Colgan, M.S.; Féret, J.-B.; Levick, S.R.; Martin, R.E.; Asner, G.P. Landscape-scale variation in plant community composition of an African savanna from airborne species mapping. Ecol. Appl. 2014, 24, 84–93. [Google Scholar] [CrossRef]
- de Cardoso, G.L.; de Araújo, G.M.; da Silva, S.A. Estrutura e dinãmica de uma população de Mauritia flexuosa L. (Arecaceae) em vereda na estação ecológica do panga, uberlândia, MG. Bol. Herbário Ezechias Paulo Heringer 2018, 9, 917820. [Google Scholar]
- Holm, J.A.; Miller, C.J.; Cropper, W.P. Population Dynamics of the Dioecious Amazonian Palm Mauritia flexuosa: Simulation Analysis of Sustainable Harvesting. Biotropica 2008, 40, 550–558. [Google Scholar] [CrossRef]
- Reynel, C.; Pennington, R.T.; Pennington, T.; Daza, A. Arboles Útiles de la Amazonía Peruana: Un Manual con Apuntes de Identificación, Ecología y Propagación de las Especies; Tarea Gráfica Educativa: Lima, Perú, 2003; ISBN 978-9972-9733-1-4. [Google Scholar]
- Ramírez-Barahona, S.; Luna-Vega, I.; Tejero-Díez, D. Species richness, endemism, and conservation of American tree ferns (Cyatheales). Biodivers. Conserv. 2011, 20, 59–72. [Google Scholar] [CrossRef]
- Ramírez-Barahona, S.; Luna-Vega, I. Geographic Differentiation of Tree Ferns (Cyatheales) in Tropical America. Am. Fern J. 2015, 105, 73–85. [Google Scholar] [CrossRef]
- Andersson, L. A Revision of the Genus Cinchona (Rubiaceae-Cinchoneae); New York Botanical Garden: New York, NY, USA, 1997; ISBN 978-0-89327-417-7. [Google Scholar]
- Griscom, B.W.; Daly, D.C.; Ashton, M.S. Floristics of bamboo-dominated stands in lowland terra-firma forests of southwestern Amazonia. J. Torrey Bot. Soc. 2007, 134, 108–125. [Google Scholar] [CrossRef]
- Thomas, E.; Alcázar Caicedo, C.; Loo, J.; Kindt, R. The distribution of the Brazil nut (Bertholletia excelsa) through time: From range contraction in glacial refugia, over human-mediated expansion, to anthropogenic climate change. Bol. Mus. Para. Emílio Goeldi Ciências Nat. 2014, 9, 267–291. [Google Scholar]
- Thomas, E.; Caicedo, C.A.; McMichael, C.H.; Corvera, R.; Loo, J. Uncovering spatial patterns in the natural and human history of Brazil nut (Bertholletia excelsa) across the Amazon Basin. J. Biogeogr. 2015, 42, 1367–1382. [Google Scholar] [CrossRef]
- ter Steege, H.; Pitman, N.C.A.; Sabatier, D.; Baraloto, C.; Salomão, R.P.; Guevara, J.E.; Phillips, O.L.; Castilho, C.V.; Magnusson, W.E.; Molino, J.-F.; et al. Hyperdominance in the Amazonian tree flora. Science 2013, 342, 1243092. [Google Scholar] [CrossRef] [Green Version]
- Levis, C.; Costa, F.R.C.; Bongers, F.; Peña-Claros, M.; Clement, C.R.; Junqueira, A.B.; Neves, E.G.; Tamanaha, E.K.; Figueiredo, F.O.G.; Salomão, R.P.; et al. Persistent effects of pre-Columbian plant domestication on Amazonian forest composition. Science 2017, 355, 925–931. [Google Scholar] [CrossRef]
- MINAM Mapa Nacional de Cobertura Vegetal (Memoria Descriptiva). Available online: http://sinia.minam.gob.pe/documentos/mapa-nacional-cobertura-vegetal-memoria-descriptiva (accessed on 15 March 2018).




| Variables | Partial Mantel Test (1) | NMDS 1 (2) | NMDS 2 (2) | NMDS 3 (2) | |
|---|---|---|---|---|---|
| Landsat | 1. Band 1 | 0.32 *** | 0.26 *** | 0 | 0.05 ** |
| 2. Band 2 | 0.42 *** | 0.38 *** | 0.01 | 0.01 | |
| 3. Band 3 | 0.39 *** | 0.3 *** | 0.01 | 0.01 | |
| 4. Band 4 | 0.38 *** | 0.45 *** | 0 | 0.01 | |
| 5. Band 5 | 0.5 *** | 0.37 *** | 0 | 0.04 * | |
| 6. Band 7 | 0.5 *** | 0.38 *** | 0 | 0.05 ** | |
| Environmental | 7. Bio1 | 0.26 *** | 0.11*** | 0.16 *** | 0 |
| 8. Bio2 | 0.09 * | 0.15 *** | 0.11 *** | 0.07 *** | |
| 9. Bio3 | −0.12 | 0 | 0.04 * | 0.13 *** | |
| 10. Bio4 | 0 | 0.02 | 0.16 *** | 0.07 *** | |
| 11. Bio5 | 0.17 ** | 0.04 * | 0.07 ** | 0.03 * | |
| 12. Bio6 | 0.26 *** | 0.17 *** | 0.21 *** | 0.02 | |
| 13. Bio7 | 0.03 | 0.11 *** | 0.11 *** | 0.1 *** | |
| 14. Bio8 | 0.22 *** | 0.08 *** | 0.14 *** | 0 | |
| 15. Bio9 | 0.27 *** | 0.16 *** | 0.16 *** | 0.01 | |
| 16. Bio10 | 0.25 *** | 0.1 *** | 0.13 *** | 0 | |
| 17. Bio11 | 0.28 *** | 0.11 *** | 0.2 *** | 0 | |
| 18. Bio12 | 0.12 * | 0.18 *** | 0.01 | 0.09 *** | |
| 19. Bio13 | 0.04 | 0.13 *** | 0.05 * | 0.01 | |
| 20. Bio14 | −0.03 | 0.12 *** | 0 | 0.17 *** | |
| 21. Bio15 | −0.04 | 0.04 * | 0.02 | 0.19 *** | |
| 22. Bio16 | 0.04 | 0.12 *** | 0.06 * | 0.01 | |
| 23. Bio17 | −0.03 | 0.11 *** | 0 | 0.17 *** | |
| 24. Bio18 | 0.03 | 0.08 *** | 0.01 | 0.08 ** | |
| 25. Bio19 | −0.07 | 0.13 *** | 0 | 0.12 *** | |
| 26. CEC | 0.17 ** | 0.2 *** | 0.15 *** | 0.01 | |
| 27. DEM | 0.28 *** | 0.18 *** | 0.03 * | 0 | |
| CV | Variable Combination | NMDS 1 | NMDS 2 | NMDS 3 | |||
|---|---|---|---|---|---|---|---|
| (R2) | RMSE | (R2) | RMSE | (R2) | RMSE | ||
| Random | Landsat | 0.531 | 0.455 | 0.219 | 0.467 | 0.199 | 0.467 |
| Environmental (Env) | 0.372 | 0.537 | 0.449 | 0.414 | 0.321 | 0.414 | |
| Landsat + Env | 0.625 | 0.422 | 0.430 | 0.404 | 0.358 | 0.404 | |
| Selected by ffs | 0.626 | 0.411 | 0.605 | 0.395 | 0.396 | 0.395 | |
| Selected variables (*) | 5,8,13,16,24 | 1,2,6,12,18,24,25,27 | 1,12,24,27 | ||||
| Spatial | Landsat | 0.392 | 0.484 | 0.045 | 0.486 | 0.043 | 0.486 |
| Environmental (Env) | 0.136 | 0.565 | 0.209 | 0.465 | 0.142 | 0.465 | |
| Landsat + Env | 0.483 | 0.437 | 0.201 | 0.431 | 0.166 | 0.431 | |
| Selected by ffs | 0.531 | 0.413 | 0.328 | 0.405 | 0.272 | 0.405 | |
| Selected variables (*) | 6,15,22,27 | 2,12,18,22,27 | 3,6,20,23 | ||||
| Class | Number of Inventory Plots | Number of Genera Associated | Genus | Indicator Value | p-Value |
|---|---|---|---|---|---|
| Class 1 | 22 | 5 | Capirona | 0.513 | 0.02 * |
| Quiina | 0.503 | 0.017 * | |||
| Bertholletia | 0.449 | 0.03 * | |||
| Pausandra | 0.44 | 0.044 * | |||
| Aiouea | 0.426 | 0.05 * | |||
| Class 2 | 7 | 1 | Maclura | 0.738 | 0.001 * |
| Class 3 | 11 | 2 | Juglans | 0.522 | 0.004 ** |
| Margaritaria | 0.426 | 0.035 * | |||
| Class 4 | 13 | 0 | -- | -- | -- |
| Class 5 | 8 | 10 | Batocarpus | 0.621 | 0.003 ** |
| Cavanillesia | 0.604 | 0.002 ** | |||
| Copaifera | 0.584 | 0.002 ** | |||
| Jacaratia | 0.58 | 0.003 ** | |||
| Myriocarpa | 0.561 | 0.005 ** | |||
| Class 6 | 7 | 6 | Cyathea | 0.703 | 0.001 * |
| Hieronyma | 0.571 | 0.017 * | |||
| Palicourea | 0.545 | 0.013 * | |||
| Chaunochiton | 0.535 | 0.01 ** | |||
| Cinchona | 0.526 | 0.008 ** | |||
| Class 7 | 43 | 2 | Senefeldera | 0.493 | 0.045 ** |
| Macoubea | 0.463 | 0.041 * | |||
| Class 8 | 33 | 0 | -- | -- | -- |
| Class 9 | 9 | 5 | Mauritia | 0.751 | 0.001 * |
| Pachira | 0.711 | 0.024 * | |||
| Ferdinandusa | 0.576 | 0.003 ** | |||
| Platycarpum | 0.471 | 0.024 * | |||
| Diospyros | 0.457 | 0.047 * | |||
| Class 10 | 4 | 22 | Fusaea | 0.7 | 0.001 * |
| Pseudobombax | 0.678 | 0.003 ** | |||
| Quararibea | 0.652 | 0.002 ** | |||
| Castilla | 0.651 | 0.001 * | |||
| Ampelocera | 0.633 | 0.004 ** |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chaves, P.P.; Zuquim, G.; Ruokolainen, K.; Van doninck, J.; Kalliola, R.; Gómez Rivero, E.; Tuomisto, H. Mapping Floristic Patterns of Trees in Peruvian Amazonia Using Remote Sensing and Machine Learning. Remote Sens. 2020, 12, 1523. https://0-doi-org.brum.beds.ac.uk/10.3390/rs12091523
Chaves PP, Zuquim G, Ruokolainen K, Van doninck J, Kalliola R, Gómez Rivero E, Tuomisto H. Mapping Floristic Patterns of Trees in Peruvian Amazonia Using Remote Sensing and Machine Learning. Remote Sensing. 2020; 12(9):1523. https://0-doi-org.brum.beds.ac.uk/10.3390/rs12091523
Chicago/Turabian StyleChaves, Pablo Pérez, Gabriela Zuquim, Kalle Ruokolainen, Jasper Van doninck, Risto Kalliola, Elvira Gómez Rivero, and Hanna Tuomisto. 2020. "Mapping Floristic Patterns of Trees in Peruvian Amazonia Using Remote Sensing and Machine Learning" Remote Sensing 12, no. 9: 1523. https://0-doi-org.brum.beds.ac.uk/10.3390/rs12091523