Crop Nitrogen Retrieval Methods for Simulated Sentinel-2 Data Using In-Field Spectrometer Data
Abstract
:1. Introduction
1.1. Nitrogen in Agro-Ecosystems
1.2. Remote Sensing of Plant Nitrogen and Biomass
1.2.1. On the Terminology of Plant Nitrogen Status
1.2.2. Remote Sensing of Crop Nitrogen
1.2.3. Remote Sensing Plant Biomass
1.2.4. Field Spectrometer for Validating Satellite Measurements
1.3. Aims of This Study
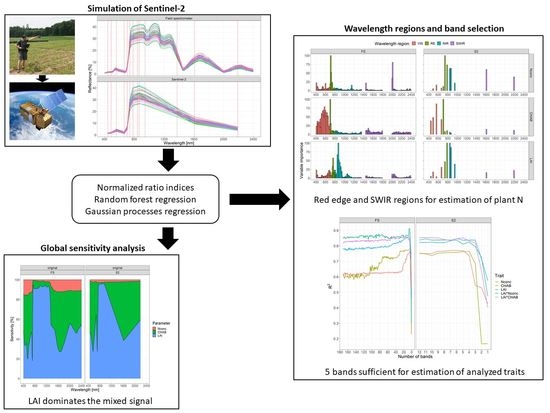
2. Materials and Methods
2.1. The Dataset
2.2. Data Analysis
2.2.1. Dataset Pre-Processing
2.2.2. Normalized Ratio Indices Generation
2.2.3. Random Forest Regression
2.2.4. Gaussian Processes Regression–Band Analysis Tool
2.2.5. Global Sensitivity Analysis
3. Results
3.1. Comparison of Spectral Analysis Methods
3.2. Spectral Band Selection
3.2.1. Random Forest Variable Importance
3.2.2. Gaussian Processes Regression–Band Analysis Tool
3.3. Global Sensitivity Analysis
4. Discussion
4.1. Optimal Analysis Method Depends on Target Trait
4.2. Low Specificity of Index-Based Methods for Satellite-Based Remote Sensing
4.3. Model Performance on Data Subsets
4.4. Influence of Band Number and Bandwidth on Trait Estimation
4.5. Spectral Regions for Trait Estimation
4.6. Important Bands for Plant N Estimation
4.7. Field Spectrometer Data for Satellite Data Simulation
4.8. Outlook on Remote Sensing of Plant N
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Muñoz-Huerta, R.; Guevara-Gonzalez, R.; Contreras-Medina, L.; Torres-Pacheco, I.; Prado-Olivarez, J.; Ocampo-Velazquez, R. A Review of Methods for Sensing the Nitrogen Status in Plants: Advantages, Disadvantages and Recent Advances. Sensors 2013, 13, 10823–10843. [Google Scholar] [CrossRef]
- Galloway, J.N.; Cowling, E.B. Reactive Nitrogen and The World: 200 Years of Change. Ambio 2002, 31, 64–71. [Google Scholar] [CrossRef]
- Chapin, F.S.; Bloom, A.J.; Field, C.B.; Waring, R.H. Plant Responses to Multiple Environmental Factors Physiological Ecology Provides Tools for Studying How Interacting Environmental Resources Control Plant Growth. BioScience 1987, 37, 49–57. [Google Scholar] [CrossRef]
- Kokaly, R.F.; Asner, G.P.; Ollinger, S.V.; Martin, M.E.; Wessman, C.A. Characterizing Canopy Biochemistry from Imaging Spectroscopy and Its Application to Ecosystem Studies. Remote Sens. Environ. 2009, 113, S78–S91. [Google Scholar] [CrossRef]
- Berger, K.; Verrelst, J.; Féret, J.-B.; Wang, Z.; Wocher, M.; Strathmann, M.; Danner, M.; Mauser, W.; Hank, T. Crop Nitrogen Monitoring: Recent Progress and Principal Developments in the Context of Imaging Spectroscopy Missions. Remote Sens. Environ. 2020, 242, 111758. [Google Scholar] [CrossRef]
- Wright, I.J.; Reich, P.B.; Westoby, M.; Ackerly, D.D.; Baruch, Z.; Bongers, F.; Cavender-Bares, J.; Chapin, T.; Cornelissen, J.H.C.; Diemer, M.; et al. The Worldwide Leaf Economics Spectrum. Nature 2004, 428, 821–827. [Google Scholar] [CrossRef]
- Haynes, R. Mineral Nitrogen in the Plant-Soil System; Elsevier: Amsterdam, The Netherlands, 2012; ISBN 978-0-323-14816-0. [Google Scholar]
- Gruber, N.; Galloway, J.N. An Earth-System Perspective of the Global Nitrogen Cycle. Nature 2008, 451, 293–296. [Google Scholar] [CrossRef] [PubMed]
- Galloway, J.N.; Townsend, A.R.; Erisman, J.W.; Bekunda, M.; Cai, Z.; Freney, J.R.; Martinelli, L.A.; Seitzinger, S.P.; Sutton, M.A. Transformation of the Nitrogen Cycle: Recent Trends, Questions, and Potential Solutions. Science 2008, 320, 889–892. [Google Scholar] [CrossRef] [Green Version]
- Diaz, R.J.; Rosenberg, R. Spreading Dead Zones and Consequences for Marine Ecosystems. Science 2008, 321, 926–929. [Google Scholar] [CrossRef] [PubMed]
- Turner, R.E.; Rabalais, N.N.; Justić, D.; Dortch, Q. Global Patterns of Dissolved N, P and Si in Large Rivers. Biogeochemistry 2003, 64, 297–317. [Google Scholar] [CrossRef]
- Dise, N.B.; Ashmore, M.; Belyazid, S.; Bleeker, A.; Bobbink, R.; de Vries, W.; Erisman, J.W.; Spranger, T.; Stevens, C.J.; van den Berg, L. Nitrogen as a threat to European terrestrial biodiversity. In The European Nitrogen Assessment; Sutton, M.A., Howard, C.M., Erisman, J.W., Billen, G., Bleeker, A., Grennfelt, P., van Grinsven, H., Grizzetti, B., Eds.; Cambridge University Press: Cambridge, MA, USA, 2011; pp. 463–494. ISBN 978-0-511-97698-8. [Google Scholar]
- Dalal, R.C.; Wang, W.; Robertson, G.P.; Parton, W.J. Nitrous Oxide Emission from Australian Agricultural Lands and Mitigation Options: A Review. Soil Res. 2003, 41, 165–195. [Google Scholar] [CrossRef]
- Wrage, N.; Velthof, G.L.; van Beusichem, M.L.; Oenema, O. Role of Nitrifier Denitrification in the Production of Nitrous Oxide. Soil Biol. Biochem. 2001, 33, 1723–1732. [Google Scholar] [CrossRef]
- Conway, G.R. Agroecosystem Analysis. Agric. Adm. 1985, 20, 31–55. [Google Scholar] [CrossRef]
- Lemaire, G.; Jeuffroy, M.-H.; Gastal, F. Diagnosis Tool for Plant and Crop N Status in Vegetative Stage. Eur. J. Agron. 2008, 28, 614–624. [Google Scholar] [CrossRef]
- Prey, L.; Schmidhalter, U. Sensitivity of Vegetation Indices for Estimating Vegetative N Status in Winter Wheat. Sensors 2019, 19, 3712. [Google Scholar] [CrossRef] [Green Version]
- Tremblay, N.; Fallon, E.; Ziadi, N. Sensing of Crop Nitrogen Status: Opportunities, Tools, Limitations, and Supporting Information Requirements. HortTecnology 2011, 21, 274–281. [Google Scholar] [CrossRef]
- Sharma, L.; Bali, S. A Review of Methods to Improve Nitrogen Use Efficiency in Agriculture. Sustainability 2017, 10, 51. [Google Scholar] [CrossRef] [Green Version]
- Argento, F.; Anken, T.; Abt, F.; Vogelsanger, E.; Walter, A.; Liebisch, F. Site-Specific Nitrogen Management in Winter Wheat Supported by Low-Altitude Remote Sensing and Soil Data. Precis. Agric. 2020. [Google Scholar] [CrossRef]
- Chen, P.; Haboudane, D.; Tremblay, N.; Wang, J.; Vigneault, P.; Li, B. New Spectral Indicator Assessing the Efficiency of Crop Nitrogen Treatment in Corn and Wheat. Remote Sens. Environ. 2010, 114, 1987–1997. [Google Scholar] [CrossRef]
- Baret, F.; Houles, V.; Guerif, M. Quantification of Plant Stress Using Remote Sensing Observations and Crop Models: The Case of Nitrogen Management. J. Exp. Bot. 2006, 58, 869–880. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jay, S.; Maupas, F.; Bendoula, R.; Gorretta, N. Retrieving LAI, Chlorophyll and Nitrogen Contents in Sugar Beet Crops from Multi-Angular Optical Remote Sensing: Comparison of Vegetation Indices and PROSAIL Inversion for Field Phenotyping. Field Crop. Res. 2017, 210, 33–46. [Google Scholar] [CrossRef] [Green Version]
- Prey, L.; Schmidhalter, U. Simulation of Satellite Reflectance Data Using High-Frequency Ground Based Hyperspectral Canopy Measurements for in-Season Estimation of Grain Yield and Grain Nitrogen Status in Winter Wheat. ISPRS J. Photogramm. Remote Sens. 2019, 149, 176–187. [Google Scholar] [CrossRef]
- Zhang, H.-Y.; Ren, X.-X.; Zhou, Y.; Wu, Y.-P.; He, L.; Heng, Y.-R.; Feng, W.; Wang, C.-Y. Remotely Assessing Photosynthetic Nitrogen Use Efficiency with in Situ Hyperspectral Remote Sensing in Winter Wheat. Eur. J. Agron. 2018, 101, 90–100. [Google Scholar] [CrossRef]
- Chang, N.-B.; Imen, S.; Vannah, B. Remote Sensing for Monitoring Surface Water Quality Status and Ecosystem State in Relation to the Nutrient Cycle: A 40-Year Perspective. Crit. Rev. Environ. Sci. Technol. 2015, 45, 101–166. [Google Scholar] [CrossRef]
- Jin, Z.; Archontoulis, S.V.; Lobell, D.B. How Much Will Precision Nitrogen Management Pay off? An Evaluation Based on Simulating Thousands of Corn Fields over the US Corn-Belt. Field Crop. Res. 2019, 240, 12–22. [Google Scholar] [CrossRef]
- Stroppiana, D.; Fava, F.; Boschetti, M.; Brivio, P. Estimation of Nitrogen Content in Crops and Pastures Using Hyperspectral Vegetation Indices. In Hyperspectral Remote Sensing of Vegetation; CRC Press: Boca Raton, FL, USA, 2011; pp. 245–262. ISBN 978-1-4398-4537-0. [Google Scholar]
- Fitzgerald, G.; Rodriguez, D.; O’Leary, G. Measuring and Predicting Canopy Nitrogen Nutrition in Wheat Using a Spectral Index—The Canopy Chlorophyll Content Index (CCCI). Field Crop. Res. 2010, 116, 318–324. [Google Scholar] [CrossRef]
- Homolová, L.; Malenovský, Z.; Clevers, J.G.P.W.; García-Santos, G.; Schaepman, M.E. Review of Optical-Based Remote Sensing for Plant Trait Mapping. Ecol. Complex. 2013, 15, 1–16. [Google Scholar] [CrossRef] [Green Version]
- Erdle, K.; Mistele, B.; Schmidhalter, U. Comparison of Active and Passive Spectral Sensors in Discriminating Biomass Parameters and Nitrogen Status in Wheat Cultivars. Field Crop. Res. 2011, 124, 74–84. [Google Scholar] [CrossRef]
- Söderström, M.; Piikki, K.; Stenberg, M.; Stadig, H.; Martinsson, J. Producing Nitrogen (N) Uptake Maps in Winter Wheat by Combining Proximal Crop Measurements with Sentinel-2 and DMC Satellite Images in a Decision Support System for Farmers. Acta Agric. Scand. Sect. B Soil Plant Sci. 2017, 67, 637–650. [Google Scholar] [CrossRef]
- Schlemmer, M.; Gitelson, A.; Schepers, J.; Ferguson, R.; Peng, Y.; Shanahan, J.; Rundquist, D. Remote Estimation of Nitrogen and Chlorophyll Contents in Maize at Leaf and Canopy Levels. Int. J. Appl. Earth Obs. Geoinf. 2013, 25, 47–54. [Google Scholar] [CrossRef] [Green Version]
- Clevers, J.G.P.W.; Gitelson, A.A. Remote Estimation of Crop and Grass Chlorophyll and Nitrogen Content Using Red-Edge Bands on Sentinel-2 and -3. Int. J. Appl. Earth Obs. Geoinf. 2013, 23, 344–351. [Google Scholar] [CrossRef]
- Herrmann, I.; Karnieli, A.; Bonfil, D.J.; Cohen, Y.; Alchanatis, V. SWIR-Based Spectral Indices for Assessing Nitrogen Content in Potato Fields. Int. J. Remote Sens. 2010, 31, 5127–5143. [Google Scholar] [CrossRef]
- Thenkabail, P.S.; Mariotto, I.; Gumma, M.K.; Middleton, E.M.; Landis, D.R.; Huemmrich, K.F. Selection of Hyperspectral Narrowbands (HNBs) and Composition of Hyperspectral Twoband Vegetation Indices (HVIs) for Biophysical Characterization and Discrimination of Crop Types Using Field Reflectance and Hyperion/EO-1 Data. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2013, 6, 427–439. [Google Scholar] [CrossRef] [Green Version]
- Yoder, B.J.; Pettigrew-Crosby, R.E. Predicting Nitrogen and Chlorophyll Content and Concentrations from Reflectance Spectra (400–2500 Nm) at Leaf and Canopy Scales. Remote Sens. Environ. 1995, 53, 199–211. [Google Scholar] [CrossRef]
- Curran, P.J. Remote Sensing of Foliar Chemistry. Remote Sens. Environ. 1989, 30, 271–278. [Google Scholar] [CrossRef]
- Tucker, C.J. Red and Photographic Infrared Linear Combinations for Monitoring Vegetation. Remote Sens. Environ. 1979, 8, 24. [Google Scholar] [CrossRef] [Green Version]
- Berntsen, J.; Thomsen, A.; Schelde, K.; Hansen, O.M.; Knudsen, L.; Broge, N.; Hougaard, H.; Hørfarter, R. Algorithms for Sensor-Based Redistribution of Nitrogen Fertilizer in Winter Wheat. Precis. Agric. 2006, 7, 65–83. [Google Scholar] [CrossRef]
- Meisinger, J.J.; Schepers, J.S.; Raun, W.R. Crop Nitrogen Requirement and Fertilization. In Agronomy Monographs; Schepers, J.S., Raun, W.R., Eds.; American Society of Agronomy, Crop Science Society of America, Soil Science Society of America: Madison, WI, USA, 2015; pp. 563–612. ISBN 978-0-89118-191-0. [Google Scholar]
- Tremblay, N.; Wang, Z.; Ma, B.-L.; Belec, C.; Vigneault, P. A Comparison of Crop Data Measured by Two Commercial Sensors for Variable-Rate Nitrogen Application. Precis. Agric. 2009, 10, 145–161. [Google Scholar] [CrossRef]
- Hansen, P.M.; Schjoerring, J.K. Reflectance Measurement of Canopy Biomass and Nitrogen Status in Wheat Crops Using Normalized Difference Vegetation Indices and Partial Least Squares Regression. Remote Sens. Environ. 2003, 86, 542–553. [Google Scholar] [CrossRef]
- Pullanagari, R.R.; Kereszturi, G.; Yule, I.J. Mapping of Macro and Micro Nutrients of Mixed Pastures Using Airborne AisaFENIX Hyperspectral Imagery. ISPRS J. Photogramm. Remote Sens. 2016, 117, 1–10. [Google Scholar] [CrossRef]
- Liang, L.; Di, L.; Huang, T.; Wang, J.; Lin, L.; Wang, L.; Yang, M. Estimation of Leaf Nitrogen Content in Wheat Using New Hyperspectral Indices and a Random Forest Regression Algorithm. Remote Sens. 2018, 10, 1940. [Google Scholar] [CrossRef] [Green Version]
- Berger, K.; Verrelst, J.; Féret, J.-B.; Hank, T.; Wocher, M.; Mauser, W.; Camps-Valls, G. Retrieval of Aboveground Crop Nitrogen Content with a Hybrid Machine Learning Method. Int. J. Appl. Earth Obs. Geoinf. 2020, 92, 102174. [Google Scholar] [CrossRef]
- Van Wittenberghe, S.; Verrelst, J.; Rivera, J.P.; Alonso, L.; Moreno, J.; Samson, R. Gaussian Processes Retrieval of Leaf Parameters from a Multi-Species Reflectance, Absorbance and Fluorescence Dataset. J. Photochem. Photobiol. B Biol. 2014, 134, 37–48. [Google Scholar] [CrossRef]
- Wang, Z.; Townsend, P.A.; Schweiger, A.K.; Couture, J.J.; Singh, A.; Hobbie, S.E.; Cavender-Bares, J. Mapping Foliar Functional Traits and Their Uncertainties across Three Years in a Grassland Experiment. Remote Sens. Environ. 2019, 221, 405–416. [Google Scholar] [CrossRef]
- Chlingaryan, A.; Sukkarieh, S.; Whelan, B. Machine Learning Approaches for Crop Yield Prediction and Nitrogen Status Estimation in Precision Agriculture: A Review. Comput. Electron. Agric. 2018, 151, 61–69. [Google Scholar] [CrossRef]
- Aasen, H.; Gnyp, M.L.; Miao, Y.; Bareth, G. Automated Hyperspectral Vegetation Index Retrieval from Multiple Correlation Matrices with HyperCor. Photogramm. Eng. Remote Sens. 2014, 80, 785–795. [Google Scholar] [CrossRef]
- Broge, N.H.; Leblanc, E. Comparing Prediction Power and Stability of Broadband and Hyperspectral Vegetation Indices for Estimation of Green Leaf Area Index and Canopy Chlorophyll Density. Remote Sens. Environ. 2001, 76, 156–172. [Google Scholar] [CrossRef]
- Gnyp, M.L.; Bareth, G.; Li, F.; Lenz-Wiedemann, V.I.S.; Koppe, W.; Miao, Y.; Hennig, S.D.; Jia, L.; Laudien, R.; Chen, X.; et al. Development and Implementation of a Multiscale Biomass Model Using Hyperspectral Vegetation Indices for Winter Wheat in the North China Plain. Int. J. Appl. Earth Obs. Geoinf. 2014, 33, 232–242. [Google Scholar] [CrossRef]
- Gnyp, M.L.; Miao, Y.; Yuan, F.; Ustin, S.L.; Yu, K.; Yao, Y.; Huang, S.; Bareth, G. Hyperspectral Canopy Sensing of Paddy Rice Aboveground Biomass at Different Growth Stages. Field Crop. Res. 2014, 155, 42–55. [Google Scholar] [CrossRef]
- Lambert, M.-J.; Traoré, P.C.S.; Blaes, X.; Baret, P.; Defourny, P. Estimating Smallholder Crops Production at Village Level from Sentinel-2 Time Series in Mali’s Cotton Belt. Remote Sens. Environ. 2018, 216, 647–657. [Google Scholar] [CrossRef]
- Bendig, J.; Yu, K.; Aasen, H.; Bolten, A.; Bennertz, S.; Broscheit, J.; Gnyp, M.L.; Bareth, G. Combining UAV-Based Plant Height from Crop Surface Models, Visible, and near Infrared Vegetation Indices for Biomass Monitoring in Barley. Int. J. Appl. Earth Obs. Geoinf. 2015, 39, 79–87. [Google Scholar] [CrossRef]
- Clevers, J.; Kooistra, L.; van den Brande, M. Using Sentinel-2 Data for Retrieving LAI and Leaf and Canopy Chlorophyll Content of a Potato Crop. Remote Sens. 2017, 9, 405. [Google Scholar] [CrossRef] [Green Version]
- Oliveira, R.A.; Näsi, R.; Niemeläinen, O.; Nyholm, L.; Alhonoja, K.; Kaivosoja, J.; Jauhiainen, L.; Viljanen, N.; Nezami, S.; Markelin, L.; et al. Machine Learning Estimators for the Quantity and Quality of Grass Swards Used for Silage Production Using Drone-Based Imaging Spectrometry and Photogrammetry. Remote Sens. Environ. 2020, 246, 111830. [Google Scholar] [CrossRef]
- Haboudane, D.; Miller, J.R.; Pattey, E.; Zarco-Tejada, P.J.; Strachan, I.B. Hyperspectral Vegetation Indices and Novel Algorithms for Predicting Green LAI of Crop Canopies: Modeling and Validation in the Context of Precision Agriculture. Remote Sens. Environ. 2004, 90, 337–352. [Google Scholar] [CrossRef]
- Crema, A.; Boschetti, M.; Nutini, F.; Cillis, D.; Casa, R. Influence of Soil Properties on Maize and Wheat Nitrogen Status Assessment from Sentinel-2 Data. Remote Sens. 2020, 12, 2175. [Google Scholar] [CrossRef]
- Delloye, C.; Weiss, M.; Defourny, P. Retrieval of the Canopy Chlorophyll Content from Sentinel-2 Spectral Bands to Estimate Nitrogen Uptake in Intensive Winter Wheat Cropping Systems. Remote Sens. Environ. 2018, 216, 245–261. [Google Scholar] [CrossRef]
- Meier, J.; Mauser, W.; Hank, T.; Bach, H. Assessments on the Impact of High-Resolution-Sensor Pixel Sizes for Common Agricultural Policy and Smart Farming Services in European Regions. Comput. Electron. Agric. 2020, 169, 105205. [Google Scholar] [CrossRef]
- Bundesamt für Statistik. Landwirtschaft Und Ernährung—Taschenstatistik 2020; Bundesamt für Statistik BFS: Bern, Switzerland, 2020. [Google Scholar]
- Clevers, J.G.P.W.; Kooistra, L. Using Hyperspectral Remote Sensing Data for Retrieving Canopy Chlorophyll and Nitrogen Content. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2012, 5, 574–583. [Google Scholar] [CrossRef]
- Frampton, W.J.; Dash, J.; Watmough, G.; Milton, E.J. Evaluating the Capabilities of Sentinel-2 for Quantitative Estimation of Biophysical Variables in Vegetation. ISPRS J. Photogramm. Remote Sens. 2013, 82, 83–92. [Google Scholar] [CrossRef] [Green Version]
- Morcillo-Pallarés, P.; Rivera-Caicedo, J.P.; Belda, S.; De Grave, C.; Burriel, H.; Moreno, J.; Verrelst, J. Quantifying the Robustness of Vegetation Indices through Global Sensitivity Analysis of Homogeneous and Forest Leaf-Canopy Radiative Transfer Models. Remote Sens. 2019, 11, 2418. [Google Scholar] [CrossRef] [Green Version]
- Myneni, R.B.; Ramakrishna, R.; Nemani, R.; Running, S.W. Estimation of Global Leaf Area Index and Absorbed Par Using Radiative Transfer Models. IEEE Trans. Geosci. Remote Sens. 1997, 35, 1380–1393. [Google Scholar] [CrossRef] [Green Version]
- Verrelst, J.; Malenovský, Z.; Van der Tol, C.; Camps-Valls, G.; Gastellu-Etchegorry, J.-P.; Lewis, P.; North, P.; Moreno, J. Quantifying Vegetation Biophysical Variables from Imaging Spectroscopy Data: A Review on Retrieval Methods. Surv. Geophys. 2019, 40, 589–629. [Google Scholar] [CrossRef] [Green Version]
- Liebisch, F.; Kung, G.; Damm, A.; Walter, A. Characterization of Crop Vitality and Resource Use Efficiency by Means of Combining Imaging Spectroscopy Based Plant Traits. In Proceedings of the 2014 6th Workshop on Hyperspectral Image and Signal Processing: Evolution in Remote Sensing (WHISPERS), Lausanne, Switzerland, 24–27 June 2014; pp. 1–4. [Google Scholar]
- Walter, A.; Khanna, R.; Lottes, P.; Stachniss, C.; Nieto, J.; Liebisch, F. Flourish—A Robotic Approach for Automation in Crop Management. In Proceedings of the 14th International Conference on Precision Agriculture, Montreal, QC, Canada, 24–27 June 2018; p. 9. [Google Scholar]
- Lancashire, P.D.; Bleiholder, H.; Boom, T.V.D.; Langelüddeke, P.; Stauss, R.; Weber, E.; Witzenberger, A. A Uniform Decimal Code for Growth Stages of Crops and Weeds. Ann. Appl. Biol. 1991, 119, 561–601. [Google Scholar] [CrossRef]
- Roth, L.; Aasen, H.; Walter, A.; Liebisch, F. Extracting Leaf Area Index Using Viewing Geometry Effects—A New Perspective on High-Resolution Unmanned Aerial System Photography. ISPRS J. Photogramm. Remote Sens. 2018, 141, 161–175. [Google Scholar] [CrossRef]
- Lichtenthaler, H.K.; Buschmann, C. Chlorophylls and Carotenoids: Measurement and Characterization by UV-VIS Spectroscopy. Curr. Protoc. Food Anal. Chem. 2001, 1, F4.3.1–F4.3.8. [Google Scholar] [CrossRef]
- Jones, H.G.; Vaughan, R.A. Remote Sensing of Vegetation: Principles, Techniques, and Applications; OUP Oxford: Oxford, UK, 2010; ISBN 978-0-19-920779-4. [Google Scholar]
- R Core Team. A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2013. [Google Scholar]
- Lehnert, L.W.; Meyer, H.; Obermeier, W.A.; Silva, B.; Regeling, B.; Bendix, J. Hyperspectral Data Analysis in R: The Hsdar Package. J. Stat. Soft. 2019, 89. [Google Scholar] [CrossRef] [Green Version]
- Li, F.; Gnyp, M.L.; Jia, L.; Miao, Y.; Yu, Z.; Koppe, W.; Bareth, G.; Chen, X.; Zhang, F. Estimating N Status of Winter Wheat Using a Handheld Spectrometer in the North China Plain. Field Crop. Res. 2008, 106, 9. [Google Scholar] [CrossRef]
- Gnyp, M.L.; Yu, K.; Aasen, H.; Yao, Y.; Huang, S.; Miao, Y.; Bareth, C.G. Analysis of Crop Reflectance for Estimating Biomass in Rice Canopies at Different Phenological Stages—Reflexionsanalyse Zur Abschätzung Der Biomasse von Reis in Unterschiedlichen Phänologischen Stadien. Photogramm. Fernerkund. Geoinf. 2013, 351–365. [Google Scholar] [CrossRef]
- Wang, L.; Chang, Q.; Yang, J.; Zhang, X.; Li, F. Estimation of Paddy Rice Leaf Area Index Using Machine Learning Methods Based on Hyperspectral Data from Multi-Year Experiments. PLoS ONE 2018, 13, e0207624. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuhn, M. Building Predictive Models in R Using the Caret Package. J. Stat. Softw. 2008, 28, 1–26. [Google Scholar] [CrossRef] [Green Version]
- Wright, M.N.; Ziegler, A. Ranger: A Fast Implementation of Random Forests for High Dimensional Data in C++ and R. J. Stat. Softw. 2017, 77, 1–17. [Google Scholar] [CrossRef] [Green Version]
- Nicodemus, K.K.; Malley, J.D.; Strobl, C.; Ziegler, A. The Behaviour of Random Forest Permutation-Based Variable Importance Measures under Predictor Correlation. BMC Bioinform. 2010, 11, 110. [Google Scholar] [CrossRef] [Green Version]
- Verrelst, J.; Romijn, E.; Kooistra, L. Mapping Vegetation Density in a Heterogeneous River Floodplain Ecosystem Using Pointable CHRIS/PROBA Data. Remote Sens. 2012, 4, 2866–2889. [Google Scholar] [CrossRef] [Green Version]
- Verrelst, J.; Rivera, J.P.; Mardashova, M.; Moreno, J. ARTMO’s Global Sensitivity Analysis (GSA) Toolbox to Quantify Driving Variables of Leaf and Canopy Radiative Transfer Models 2015. In Proceedings of the 9th EARSeL SIG Imaging Spectroscopy Workshop, Luxembourg, 14–16 April 2015. [Google Scholar]
- Caicedo, J.P.R.; Verrelst, J.; Munoz-Mari, J.; Moreno, J.; Camps-Valls, G. Toward a Semiautomatic Machine Learning Retrieval of Biophysical Parameters. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2014, 7, 1249–1259. [Google Scholar] [CrossRef]
- Rivera, J.P.; Verrelst, J.; Gómez-Dans, J.; Muñoz-Marí, J.; Moreno, J.; Camps-Valls, G. An Emulator Toolbox to Approximate Radiative Transfer Models with Statistical Learning. Remote Sens. 2015, 7, 9347–9370. [Google Scholar] [CrossRef] [Green Version]
- Verrelst, J.; Rivera, J.P.; Gitelson, A.; Delegido, J.; Moreno, J.; Camps-Valls, G. Spectral Band Selection for Vegetation Properties Retrieval Using Gaussian Processes Regression. Int. J. Appl. Earth Obs. Geoinf. 2016, 52, 554–567. [Google Scholar] [CrossRef]
- Rasmussen, C.E.; Williams, C.K.I. Gaussian Processes for Machine Learning; Adaptive Computation and Machine Learning Series; MIT Press: Cambridge, MA, USA, 2006; ISBN 978-0-262-18253-9. [Google Scholar]
- Verrelst, J.; Vicent, J.; Rivera-Caicedo, J.P.; Lumbierres, M.; Morcillo-Pallarés, P.; Moreno, J. Global Sensitivity Analysis of Leaf-Canopy-Atmosphere RTMs: Implications for Biophysical Variables Retrieval from Top-of-Atmosphere Radiance Data. Remote Sens. 2019, 11, 1923. [Google Scholar] [CrossRef] [Green Version]
- Camino, C.; González-Dugo, V.; Hernández, P.; Sillero, J.C.; Zarco-Tejada, P.J. Improved Nitrogen Retrievals with Airborne-Derived Fluorescence and Plant Traits Quantified from VNIR-SWIR Hyperspectral Imagery in the Context of Precision Agriculture. Int. J. Appl. Earth Obs. Geoinf. 2018, 70, 105–117. [Google Scholar] [CrossRef]
- Jay, S.; Gorretta, N.; Morel, J.; Maupas, F.; Bendoula, R.; Rabatel, G.; Dutartre, D.; Comar, A.; Baret, F. Estimating Leaf Chlorophyll Content in Sugar Beet Canopies Using Millimeter- to Centimeter-Scale Reflectance Imagery. Remote Sens. Environ. 2017, 198, 173–186. [Google Scholar] [CrossRef]
- Zhou, K.; Cheng, T.; Zhu, Y.; Cao, W.; Ustin, S.L.; Zheng, H.; Yao, X.; Tian, Y. Assessing the Impact of Spatial Resolution on the Estimation of Leaf Nitrogen Concentration Over the Full Season of Paddy Rice Using Near-Surface Imaging Spectroscopy Data. Front. Plant Sci. 2018, 9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thenkabail, P.S. Optimal Hyperspectral Narrowbands for Discriminating Agricultural Crops. Remote Sens. Rev. 2001, 20, 257–291. [Google Scholar] [CrossRef]
- Daughtry, C.S.T.; Walthall, C.L.; Kim, M.S.; de Colstoun, E.B.; McMurtrey, J.E. Estimating Corn Leaf Chlorophyll Concentration from Leaf and Canopy Reflectance. Remote Sens. Environ. 2000, 74, 229–239. [Google Scholar] [CrossRef]
- Yuan, H.; Yang, G.; Li, C.; Wang, Y.; Liu, J.; Yu, H.; Feng, H.; Xu, B.; Zhao, X.; Yang, X. Retrieving Soybean Leaf Area Index from Unmanned Aerial Vehicle Hyperspectral Remote Sensing: Analysis of RF, ANN, and SVM Regression Models. Remote Sens. 2017, 9, 309. [Google Scholar] [CrossRef] [Green Version]
- Verrelst, J.; Pablo Rivera, J.; Moreno, J.; Camps-Valls, G. Gaussian Processes Uncertainty Estimates in Experimental Sentinel-2 LAI and Leaf Chlorophyll Content Retrieval. ISPRS J. Photogramm. Remote Sens. 2013, 86, 157–167. [Google Scholar] [CrossRef]
- He, L.; Song, X.; Feng, W.; Guo, B.-B.; Zhang, Y.-S.; Wang, Y.-H.; Wang, C.-Y.; Guo, T.-C. Improved Remote Sensing of Leaf Nitrogen Concentration in Winter Wheat Using Multi-Angular Hyperspectral Data. Remote Sens. Environ. 2016, 174, 122–133. [Google Scholar] [CrossRef]
- Ziegler, A.; König, I.R. Mining Data with Random Forests: Current Options for Real-World Applications. Wires Data Min. Knowl. Discov. 2014, 4, 55–63. [Google Scholar] [CrossRef]
- Jordan, M.I.; Mitchell, T.M. Machine Learning: Trends, Perspectives, and Prospects. Science 2015, 349, 255–260. [Google Scholar] [CrossRef]
- Mousivand, A.; Menenti, M.; Gorte, B.; Verhoef, W. Global Sensitivity Analysis of the Spectral Radiance of a Soil–Vegetation System. Remote Sens. Environ. 2014, 145, 131–144. [Google Scholar] [CrossRef]
- Guo, B.-B.; Qi, S.-L.; Heng, Y.-R.; Duan, J.-Z.; Zhang, H.-Y.; Wu, Y.-P.; Feng, W.; Xie, Y.-X.; Zhu, Y.-J. Remotely Assessing Leaf N Uptake in Winter Wheat Based on Canopy Hyperspectral Red-Edge Absorption. Eur. J. Agron. 2017, 82, 113–124. [Google Scholar] [CrossRef]
- Moreno-Martínez, Á.; Camps-Valls, G.; Kattge, J.; Robinson, N.; Reichstein, M.; van Bodegom, P.; Kramer, K.; Cornelissen, J.H.C.; Reich, P.; Bahn, M.; et al. A Methodology to Derive Global Maps of Leaf Traits Using Remote Sensing and Climate Data. Remote Sens. Environ. 2018, 218, 69–88. [Google Scholar] [CrossRef] [Green Version]
- Zhao, H.; Song, X.; Yang, G.; Li, Z.; Zhang, D.; Feng, H. Monitoring of Nitrogen and Grain Protein Content in Winter Wheat Based on Sentinel-2A Data. Remote Sens. 2019, 11, 1724. [Google Scholar] [CrossRef] [Green Version]
- European Comission. Working with Parliament and Council to Make the CAP Reform Fit for the European Green Deal; European Union: Brussels, Belgium, 2020. [Google Scholar]
- Zarco-Tejada, P.J.; Miller, J.R.; Harron, J.; Hu, B.; Noland, T.L.; Goel, N.; Mohammed, G.H.; Sampson, P. Needle Chlorophyll Content Estimation through Model Inversion Using Hyperspectral Data from Boreal Conifer Forest Canopies. Remote Sens. Environ. 2004, 89, 189–199. [Google Scholar] [CrossRef]
- Gnyp, M.L.; Panitzki, M.; Reusch, S.; Jasper, J.; Bolten, A.; Bareth, G. Comparison between Tractor-Based and UAV-Based Spectrometer Measurements in Winter Wheat. In Proceedings of the 13th International Conference on Precision Agriculture, St. Louis, MO, USA, 31 July–3 August 2016; p. 10. [Google Scholar]





| Individual Traits | n | Min | Median | Max |
|---|---|---|---|---|
| Nconc [%] | 322 | 0.68 | 3.46 | 5.32 |
| ChlAB [mg g−1] | 194 | 2.28 | 5.34 | 7.11 |
| LAI [m2 m−2] | 272 | 0.05 | 2.09 | 8.63 |
| LAI*Nconc [%] | 210 | 0.17 | 7.20 | 41.25 |
| LAI*ChlAB [mg g−1] | 193 | 0.14 | 11.25 | 56.01 |
| Combined Data | n | Min BBCH | Median BBCH | Max BBCH |
| full dataset | 180 | 15 | 30 | 80 |
| erectophile | 98 | 15 | 31 | 80 |
| planophile | 55 | 15 | 22 | 67 |
| winter wheat | 64 | 15 | 30 | 32 |
| sugar beet | 45 | 15 | 21 | 38 |
| Band | Band Name | Center Wavelength [nm] | Bandwidth [nm] | Ground Resolution [m] |
|---|---|---|---|---|
| B01 | Coastal aerosol | 443 | 21.00 | 60 |
| B02 | Blue | 490 | 66.00 | 10 |
| B03 | Green | 560 | 36.00 | 10 |
| B04 | Red | 665 | 31.00 | 10 |
| B05 | RE1 | 705 | 15.50 | 20 |
| B06 | RE2 | 740 | 15.00 | 20 |
| B07 | RE3 | 783 | 20.00 | 20 |
| B08 | NIR1 | 842 | 106.00 | 10 |
| B8a | NIR2 | 865 | 21.50 | 20 |
| B09 | Water vapour | 945 | 20.50 | 60 |
| B10 | SWIR—cirrus | 1375 | 30.50 | 60 |
| B11 | SWIR1 | 1610 | 92.50 | 20 |
| B12 | SWIR2 | 2190 | 180.00 | 20 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Perich, G.; Aasen, H.; Verrelst, J.; Argento, F.; Walter, A.; Liebisch, F. Crop Nitrogen Retrieval Methods for Simulated Sentinel-2 Data Using In-Field Spectrometer Data. Remote Sens. 2021, 13, 2404. https://0-doi-org.brum.beds.ac.uk/10.3390/rs13122404
Perich G, Aasen H, Verrelst J, Argento F, Walter A, Liebisch F. Crop Nitrogen Retrieval Methods for Simulated Sentinel-2 Data Using In-Field Spectrometer Data. Remote Sensing. 2021; 13(12):2404. https://0-doi-org.brum.beds.ac.uk/10.3390/rs13122404
Chicago/Turabian StylePerich, Gregor, Helge Aasen, Jochem Verrelst, Francesco Argento, Achim Walter, and Frank Liebisch. 2021. "Crop Nitrogen Retrieval Methods for Simulated Sentinel-2 Data Using In-Field Spectrometer Data" Remote Sensing 13, no. 12: 2404. https://0-doi-org.brum.beds.ac.uk/10.3390/rs13122404