Classification of Soybean Genotypes Assessed Under Different Water Availability and at Different Phenological Stages Using Leaf-Based Hyperspectral Reflectance
Abstract
:1. Introduction
2. Materials and Methods
2.1. Experimental Setup
2.1.1. Experiment I: 2016/2017, 2017/2018, and 2018/2019 cropping seasons
2.1.2. Experiment II: Unoeste University Experiment
2.2. Weather Monitoring
2.3. Spectral Data Acquisition and Processing
- 1)
- Embrapa 2016/2017 was comprised of spectral data of genotypes 1, 2, 3, and 4 collected in 29, 34, 45, 58, 70, 90, and 113 days after sowing (DAS) in the four experimental treatments during the 2016/2017 cropping season at Embrapa Soja, with four subsamples in each plot, resulting in 1776 leaf reflectance spectra, totaling 444 spectral samples.
- 2)
- Embrapa 2017/2018 was comprised of the spectral data of genotypes 1, 2, 3, 4, and 5 collected in 30, 38, 43, 58, 79, 97, 107, and 113 DAS in the four experimental treatments during the 2017/2018 cropping season at Embrapa Soja, with four subsamples in each plot, resulting in 1920 leaf reflectance spectra, generating 640 spectral samples.
- 3)
- Embrapa 2018/2019 [I] was comprised of spectral data of genotypes 1, 2, 3, 4, and 5 collected in 42, 51, 58, 65, 80, 88, 95, 102, and 108 DAS in the ‘non-irrigated’ treatment during the 2018/2019 cropping season at Embrapa Soja, totaling 900 spectral samples.
- 4)
- Embrapa 2018/2019 [II] was comprised of spectral data of genotypes 6, 7, 8, 9, and 10 collected in 42, 51, 58, 65, 80, 88, 95, 102, and 108 DAS in the four experimental treatments during the 2018/2019 cropping season at Embrapa Soja, with four subsamples in each plot, resulting in 2880 leaf reflectance spectra and 720 spectral samples.
- 5)
- Embrapa 2016–2019 was comprised of the ‘Embrapa 2016/2017’, ‘Embrapa 2017/2018’, and ‘Embrapa 2018/2019 [I]’ spectral datasets, adding up to 1984 spectral samples of genotypes 1, 2, 3, 4, and 5.
- 6)
- Unoeste 2018/2019 [I] was comprised of spectral data of genotypes 1, 2, 3, 4, and 5 collected in 51, 62, 92, and 107 DAS during the 2018/2019 cropping season at the Unoeste University farm, resulting in 525 spectral samples.
- 7)
- Unoeste 2018/2019 [II] was comprised of spectral data of genotypes 6, 8, 9, and 10 collected at 51, 62, 92, and 107 DAS during the 2018/2019 cropping season at the Unoeste University farm, totaling 420 spectral samples.
- 8)
- Embrapa 2016–2019 + Unoeste [I] was comprised of the ‘Embrapa 2016–2019’ and ‘Unoeste 2018/2019 [I]’ spectral datasets, for a total of 2509 spectral samples of genotypes 1, 2, 3, 4, and 5.
- 9)
- Embrapa 2018/2019 + Unoeste [II] was comprised of the ‘Embrapa 2018/2019 [II]’ and ‘Unoeste 2018/2019 [II]’ spectral datasets, summing up to 1140 spectral samples of genotypes 6, 7, 8, 9, and 10.
2.4. Statistical Analysis
2.4.1. Principal Component Analysis (PCA)
2.4.2. Linear Discriminant Analysis (LDA)
3. Results and Discussion
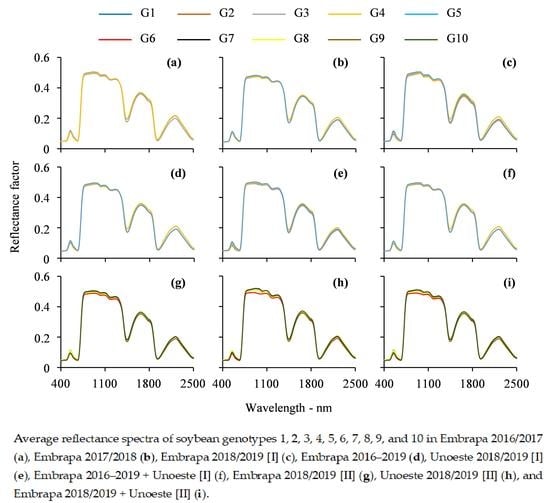
3.1. Visual Analysis of Leaf Reflectance Spectra
3.2. Principal Component Analysis—PCA
3.3. Stepwise Procedure
3.4. Linear Discriminant Analysis–LDA
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- CONAB (National Company of Food Supply). Brazilian Crop Assessment–Grain, 2019/2020 Crops, Sixth Inventory Survey, March/2020. 2020. Available online: https://www.conab.gov.br/info-agro/safras/graos/boletim-da-safra-de-graos (accessed on 25 March 2020).
- USDA (United States Department of Agriculture). World Agricultural Production. Circular Series WAP 3-20, March 2020. 2020. Available online: https://apps.fas.usda.gov/psdonline/circulars/production.pdf (accessed on 25 March 2020).
- Gusso, A.; Ducati, J.R. Algorithm for soybean classification using medium resolution satellite images. Remote Sens. 2012, 4, 3127–3142. [Google Scholar] [CrossRef] [Green Version]
- Silva Junior, C.A.; da Nanni, M.R.; Shakir, M.; Teodoro, P.E.; de Oliveira-Júnior, J.F.; Cezar, E.; Gois, G.; de Lima, M.; Wojciechowski, J.C.; Shiratsuchi, L.S. Soybean varieties discrimination using non-imaging hyperspectral sensor. Infrared Phys. Technol. 2018, 89, 338–350. [Google Scholar] [CrossRef]
- Song, X.P.; Potapov, P.V.; Krylov, A.; King, L.; Di Bella, C.M.; Hudson, A.; Khan, A.; Adusei, B.; Stehman, S.V.; Hansen, M.C. National-scale soybean mapping and area estimation in the United States using medium resolution satellite imagery and field survey. Remote Sens. Environ. 2017, 190, 383–395. [Google Scholar] [CrossRef]
- Souza, C.H.W.; de Mercante, E.; Johann, J.A.; Lamparelli, R.A.C.; Uribe-Opazo, M.A. Mapping and discrimination of soya bean and corn crops using spectro-temporal profiles of vegetation indices. Int. J. Remote Sens. 2015, 36, 1809–1824. [Google Scholar] [CrossRef]
- Sentelhas, P.C.; Battisti, R.; Câmara, G.M.S.; Farias, J.R.B.; Hampf, A.C.; Nendel, C. The soybean yield gap in Brazil–magnitude, causes and possible solutions for sustainable production. J. Agric. Sci. 2015, 153, 1394–1411. [Google Scholar] [CrossRef] [Green Version]
- Ferreira, R.C. Quantificação das Perdas por seca na Cultura da soja o Brasil. Ph.D. Thesis, Universidade Estadual de Londrina, Londrina, Brazil, 2016. [Google Scholar]
- Fuganti-Pagliarini, R.; Ferreira, L.C.; Rodrigues, F.A.; Molinari, H.B.; Marin, S.R.; Molinari, M.D.C.; Marcolino-Gomes, J.; Mertz-Henning, L.M.; Farias, J.R.B.; Oliveira, M.C.N.; et al. Characterization of soybean genetically modified for drought tolerance in field conditions. Front. Plant Sci. 2017, 8, 448. [Google Scholar] [CrossRef] [Green Version]
- Marinho, J.P.; Kanamori, N.; Ferreira, L.C.; Fuganti-Pagliarini, R.; Carvalho, J.D.F.C.; Freitas, R.A.; Marin, S.R.R.; Rodrigues, F.A.; Mertz-Henning, L.M.; Farias, J.R.B.; et al. Characterization of molecular and physiological responses under water deficit of genetically modified soybean plants overexpressing the AtAREB1 transcription factor. Plant Mol. Biol. Rep. 2016, 34, 410–426. [Google Scholar] [CrossRef]
- Buttel, F.H.; Belsky, J. Biotechnology, plant breeding, and intellectual property: Social and ethical dimensions. Sci. Technol. Hum. Values 1987, 12, 31–49. [Google Scholar] [CrossRef]
- Goldsmith, P.; Ramos, G.; Steiger, C. Intellectual property piracy in a North–South context: Empirical evidence. Agric. Econ. 2006, 35, 335–349. [Google Scholar] [CrossRef]
- Jing, H.U. Prevention and Management of Bio-Piracy of Genetic Resources from the Perspective of Intellectual Property. In Proceedings of the DEStech Transactions on Social Science, Education and Human Science (icaem), Qingdao, China, 16–17 December 2017. [Google Scholar]
- Mascarenhas, M.; Busch, L. Seeds of change: Intellectual property rights, genetically modified soybeans and seed saving in the US. Sociol. Rural. 2006, 46, 122–138. [Google Scholar] [CrossRef]
- Schnepf, R. Genetically engineered soybeans: Acceptance and intellectual property rights issues in South America. In Congressional Research Service, the Library of Congress. Resources, Science, and Industry Division; Congressional Research Service: Washington, DC, USA, 2003. [Google Scholar]
- Stein, H. Intellectual property and genetically modified seeds: The United States, trade, and the developing world. Northwestern J. Technol. Intellect. Prop. 2005, 3, 151. [Google Scholar]
- Williams Junior, S.B. Protection of plant varieties and parts as intellectual property. Science 1984, 225, 18–23. [Google Scholar] [CrossRef] [PubMed]
- Chaves, M.; de Carvalho Alves, M.; de Oliveira, M.; Sáfadi, T. A Geostatistical Approach for Modeling Soybean Crop Area and Yield Based on Census and Remote Sensing Data. Remote Sens. 2018, 10, 680. [Google Scholar] [CrossRef] [Green Version]
- da Silva Junior, C.A.; Nanni, M.R.; Teodoro, P.E.; Silva, G.F.C. Vegetation indices for discrimination of soybean areas: A new approach. Agron. J. 2017, 109, 1331–1343. [Google Scholar] [CrossRef]
- Zhong, L.; Hu, L.; Yu, L.; Gong, P.; Biging, G.S. Automated mapping of soybean and corn using phenology. ISPRS J. Photogramm. Remote Sens. 2016, 119, 151–164. [Google Scholar] [CrossRef] [Green Version]
- Herrmann, I.; Vosberg, S.; Ravindran, P.; Singh, A.; Chang, H.X.; Chilvers, M.; Conley, S.P.; Townsend, P. Leaf and canopy level detection of Fusarium virguliforme(sudden death syndrome) in soybean. Remote Sens. 2018, 10, 426. [Google Scholar] [CrossRef] [Green Version]
- Mahlein, A.K.; Rumpf, T.; Welke, P.; Dehne, H.W.; Plümer, L.; Steiner, U.; Oerke, E.C. Development of spectral indices for detecting and identifying plant diseases. Remote Sens. Environ. 2013, 128, 21–30. [Google Scholar] [CrossRef]
- Fletcher, R.S.; Reddy, K.N. Random forest and leaf multispectral reflectance data to differentiate three soybean varieties from two pigweeds. Comput. Electron. Agric. 2016, 128, 199–206. [Google Scholar] [CrossRef]
- Samseemoung, G.; Soni, P.; Jayasuriya, H.P.; Salokhe, V.M. Application of low altitude remote sensing(LARS) platform for monitoring crop growth and weed infestation in a soybean plantation. Precis. Agric. 2012, 13, 611–627. [Google Scholar] [CrossRef]
- Adams, M.L.; Norvell, W.A.; Philpot, W.D.; Peverly, J.H. Spectral detection of micronutrient deficiency in ‘Bragg’ soybean. Agron. J. 2000, 92, 261–268. [Google Scholar]
- Adams, M.L.; Norvell, W.A.; Philpot, W.D.; Peverly, J.H. Toward the discrimination of manganese, zinc, copper, and iron deficiency in ‘Bragg’ soybean using spectral detection methods. Agron. J. 2000, 92, 268–274. [Google Scholar]
- Furlanetto, R.H. Sensores Multi e Hiperespectrais na Identificação e Quantificação da Deficiência de Potássio na Cultura do Milho (Zea mays). Master’s Thesis, Universidade Estadual de Maringá, Maringá, Brazil, 2018. [Google Scholar]
- Milton, N.M.; Eiswerth, B.A.; Ager, C.M. Effect of phosphorus deficiency on spectral reflectance and morphology of soybean plants. Remote Sens. Environ. 1991, 36, 121–127. [Google Scholar] [CrossRef]
- Crusiol, L.G.T.; Carvalho, J.D.F.C.; Sibaldelli, R.N.R.; Neiverth, W.; do Rio, A.; Ferreira, L.C.; de Procópio, S.O.; Mertz-Henning, L.M.; Nepomuceno, A.L.; Neumaier, N.; et al. NDVI variation according to the time of measurement, sampling size, positioning of sensor and water regime in different soybean cultivars. Precis. Agric. 2017, 18, 470–490. [Google Scholar] [CrossRef] [Green Version]
- Maimaitiyiming, M.; Ghulam, A.; Bozzolo, A.; Wilkins, J.L.; Kwasniewski, M.T. Early detection of plant physiological responses to different levels of water stress using reflectance spectroscopy. Remote Sens. 2017, 9, 745. [Google Scholar] [CrossRef] [Green Version]
- Sakamoto, T.; Wardlow, B.D.; Gitelson, A.A.; Verma, S.B.; Suyker, A.E.; Arkebauer, T.J. A two-step filtering approach for detecting maize and soybean phenology with time-series MODIS data. Remote Sens. Environ. 2010, 114, 2146–2159. [Google Scholar] [CrossRef]
- Bolton, D.K.; Friedl, M.A. Forecasting crop yield using remotely sensed vegetation indices and crop phenology metrics. Agric. For. Meteorol. 2013, 173, 74–84. [Google Scholar] [CrossRef]
- Johnson, D.M. An assessment of pre-and within-season remotely sensed variables for forecasting corn and soybean yields in the United States. Remote Sens. Environ. 2014, 141, 116–128. [Google Scholar] [CrossRef]
- Yu, N.; Li, L.; Schmitz, N.; Tian, L.F.; Greenberg, J.A.; Diers, B.W. Development of methods to improve soybean yield estimation and predict plant maturity with an unmanned aerial vehicle based platform. Remote Sens. Environ. 2016, 187, 91–101. [Google Scholar] [CrossRef]
- Aouidi, F.; Dupuy, N.; Artaud, J.; Roussos, S.; Msallem, M.; Perraud-Gaime, I.; Hamdi, M. Discrimination of five Tunisian cultivars by Mid InfraRed spectroscopy combined with chemometric analyses of olive Olea europaea leaves. Food Chem. 2012, 131, 360–366. [Google Scholar] [CrossRef]
- Diago, M.P.; Fernandes, A.M.; Millan, B.; Tardáguila, J.; Melo-Pinto, P. Identification of grapevine varieties using leaf spectroscopy and partial least squares. Comput. Electron. Agric. 2013, 99, 7–13. [Google Scholar] [CrossRef]
- Maimaitiyiming, M.; Miller, A.J.; Ghulam, A. Discriminating spectral signatures among and within two closely related grapevine species. Photogramm. Eng. Remote Sens. 2016, 82, 51–62. [Google Scholar] [CrossRef] [Green Version]
- Gutiérrez, S.; Tardaguila, J.; Fernández-Novales, J.; Diago, M.P. Support vector machine and artificial neural network models for the classification of grapevine varieties using a portable NIR spectrophotometer. PLoS ONE 2015, 10, e0143197. [Google Scholar] [CrossRef] [PubMed]
- Sinha, P.; Robson, A.; Schneider, D.; Kilic, T.; Mugera, H.K.; Ilukor, J.; Tindamanyire, J.M. The potential of in-situ hyperspectral remote sensing for differentiating 12 banana genotypes grown in Uganda. ISPRS J. Photogramm. Remote Sens. 2020, 167, 85–103. [Google Scholar] [CrossRef]
- Lin, W.S.; Yang, C.M.; Kuo, B.J. Classifying cultivars of rice(Oryza sativa L.) based on corrected canopy reflectance spectra data using the orthogonal projections to latent structures(O-PLS) method. Chemom. Intell. Lab. Syst. 2012, 115, 25–36. [Google Scholar] [CrossRef]
- Ajayi, S.; Reddy, S.K.; Gowda, P.H.; Xue, Q.; Rudd, J.C.; Pradhan, G.; Liu, B.A.; Stewart, C.; Biradar, K.E.; Jessup, K.E. Spectral reflectance models for characterizing winter wheat genotypes. J. Crop. Improv. 2016, 30, 176–195. [Google Scholar] [CrossRef]
- Garriga, M.; Romero-Bravo, S.; Estrada, F.; Escobar, A.; Matus, I.A.; del Pozo, A.; Astudillo, C.A.; Lobos, G.A. Assessing wheat traits by spectral reflectance: Do we really need to focus on predicted trait-values or directly identify the elite genotypes group? Front. Plant Sci. 2017, 8, 280. [Google Scholar] [CrossRef] [Green Version]
- Breunig, F.M.; Galvao, L.S.; Formaggio, A.R.; Epiphanio, J.C. Classification of soybean varieties using different techniques: Case study with Hyperion and sensor spectral resolution simulations. J. Appl. Remote Sens. 2011, 5, 053533. [Google Scholar] [CrossRef] [Green Version]
- Galvão, L.S.; Roberts, D.A.; Formaggio, A.R.; Numata, I.; Breunig, F.M. View angle effects on the discrimination of soybean varieties and on the relationships between vegetation indices and yield using off-nadir Hyperion data. Remote Sens. Environ. 2009, 113, 846–856. [Google Scholar] [CrossRef]
- Ghulam, A.; Fishman, J.; Maimaitiyiming, M. Spectral separability analysis of five soybean cultivars with different ozone tolerance using hyperspectral field spectroscopy. In Proceedings of the 2016 IEEE International Geoscience and Remote Sensing Symposium (IGARSS) (6312–6315), Beijing, China, 10–15 July 2016. [Google Scholar]
- Wrege, M.S.; Steinmetz, S.; Reiser Júnior, C.; de Almeida, I.R. Atlas climático da Região Sul do Brasil: Estados do Paraná, Santa Catarina e Rio Grande do Sul; Pelotas: Embrapa Clima Temperado, Colombo: Embrapa Florestas, Brazil, 2011. [Google Scholar]
- Alvares, C.A.; Stape, J.L.; Sentelhas, P.C.; de Moraes Gonçalves, J.L.; Sparovek, G. Köppen’s climate classification map for Brazil. Meteorol. Z. 2013, 22, 711–728. [Google Scholar] [CrossRef]
- USDA (United States Department of Agriculture)—Natural Resources Conservation Service. Soil Taxonomy: A Basic System of Soil Classification for Making and Interpreting Soil Surveys; USDA: Washington, DC, USA, 1999.
- Kaster, M.; Farias, J.R.B. Regionalização dos Testes de Valor de Cultivo e Uso e da Indicação de Cultivares de Soja-Terceira Aproximação; Embrapa Soja-Documentos, 2012; Londrina: Embrapa Soja, Brazil, 2012. [Google Scholar]
- Embrapa Soja. Tecnologias de Produção de Soja–Região Central do Brasil 2014; Londrina: Embrapa Soja, Brazil, 2013. [Google Scholar]
- Sibaldelli, R.N.R.; Farias, J.R.B. Boletim Agrometeorológico da Embrapa Soja, Londrina, PR–2016; Londrina: Embrapa Soja, Brazil, 2017; Available online: http://www.infoteca.cnptia.embrapa.br/infoteca/handle/doc/1067152 (accessed on 15 June 2020).
- Sibaldelli, R.N.R.; Farias, J.R.B. Boletim Agrometeorológico da Embrapa Soja, Londrina, PR–2017; Londrina: Embrapa Soja, Brazil, 2018; Available online: https://www.infoteca.cnptia.embrapa.br/infoteca/handle/doc/1087963 (accessed on 15 June 2020).
- Sibaldelli, R.N.R.; Farias, J.R.B. Boletim Agrometeorológico da Embrapa Soja, Londrina, PR–2018; Londrina: Embrapa Soja, Brazil, 2019; Available online: https://www.infoteca.cnptia.embrapa.br/infoteca/bitstream/doc/1109091/1/DOC4111.pdf (accessed on 15 June 2020).
- Thornthwaite, C.W.; Mather, J.R. The Water Balance; Laboratory of Climatology: Centerton, AR, USA, 1955. [Google Scholar]
- Rumpf, T.; Mahlein, A.K.; Steiner, U.; Oerke, E.C.; Dehne, H.W.; Plümer, L. Early detection and classification of plant diseases with support vector machines based on hyperspectral reflectance. Comput. Electron. Agric. 2010, 74, 91–99. [Google Scholar] [CrossRef]
- Mahlein, A.K.; Steiner, U.; Dehne, H.W.; Oerke, E.C. Spectral signatures of sugar beet leaves for the detection and differentiation of diseases. Precis. Agric. 2010, 11, 413–431. [Google Scholar] [CrossRef]
- Streher, A.S.; da Silva Torres, R.; Morellato, L.P.C.; Silva, T.S.F. Accuracy and limitations for spectroscopic prediction of leaf traits in seasonally dry tropical environments. Remote Sens. Environ. 2020, 244, 111828. [Google Scholar] [CrossRef]
- Peng, Y.; Fan, M.; Song, J.; Cui, T.; Li, R. Assessment of plant species diversity based on hyperspectral indices at a fine scale. Sci. Rep. 2018, 8, 4776. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schmidt, K.S.; Skidmore, A.K. Spectral discrimination of vegetation types in a coastal wetland. Remote Sens. Environ. 2003, 85, 92–108. [Google Scholar] [CrossRef]
- Jolliffe, I.T.; Cadima, J. Principal component analysis: A review and recent developments. Philos. Trans. R. Soc. A Math. Phys. Eng. Sci. 2016, 374, 20150202. [Google Scholar] [CrossRef]
- Li, X.; He, Y.; Fang, H. Non-destructive discrimination of Chinese bayberry varieties using Vis/NIR spectroscopy. J. Food Eng. 2007, 81, 357–363. [Google Scholar] [CrossRef]
- Wang, H.W. Partial Least Squares Regression Method and Applications; National Defense Industry Press: Beijing, China, 1999; pp. 1–274. [Google Scholar]
- Karimi, Y.; Prasher, S.O.; Mcnairn, H.; Bonnell, R.B.; Dutilleul, P.; Goel, P.K. Classification accuracy of discriminant analysis, artificial neural networks, and decision trees for weed and nitrogen stress detection in corn. Trans. ASAE 2005, 48, 261–1268. [Google Scholar] [CrossRef]
- Thenkabail, P.S.; Enclona, E.A.; Ashton, M.S.; Van Der Meer, B. Accuracy assessments of hyperspectral waveband performance for vegetation analysis applications. Remote Sens. Environ. 2004, 91, 354–376. [Google Scholar] [CrossRef]
- Draper, N.R.; Smith, H. Applied Regression Analysis, 3rd ed.; John Wiley Sons: Hoboken, NJ, USA, 2014. [Google Scholar]
- Falcioni, R.; Moriwaki, T.; Bonato, C.M.; Souza, L.A.; de Nanni, M.R.; Antunes, W.C. Distinct growth light and gibberellin regimes alter leaf anatomy and reveal their influence on leaf optical properties. Environ. Exp. Bot. 2017, 140, 86–95. [Google Scholar] [CrossRef]
- Falcioni, R.; Moriwaki, T.; Pattaro, M.; Furlanetto, R.H.; Nanni, M.R.; Antunes, W.C. High resolution leaf spectral signature as a tool for foliar pigment estimation displaying potential for species differentiation. J. Plant Physiol. 2020, 249, 153161. [Google Scholar] [CrossRef]
- Moriwaki, T.; Falcioni, R.; Tanaka, F.A.O.; Cardoso, K.A.K.; Souza, L.A.; Benedito, E.; Nanni, M.R.; Bonato, C.M.; Antunes, W.C. Nitrogen-improved photosynthesis quantum yield is driven by increased thylakoid density, enhancing green light absorption. Plant Sci. 2019, 278, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Nanni, M.R.; Demattê, J.A.M. Spectral reflectance methodology in comparison to traditional soil analysis. Soil Sci. Soc. Am. J. 2006, 70, 393–407. [Google Scholar] [CrossRef]
- Price, J.C. How unique are spectral signatures? Remote Sens. Environ. 1994, 49, 181–186. [Google Scholar] [CrossRef]
- Breunig, F.M.; Galvão, L.S.; Formaggio, A.R.; Epiphanio, J.C. Variation of MODIS reflectance and vegetation indices with viewing geometry and soybean development. An. Acad. Bras. Ciênc. 2012, 84, 263–274. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Crusiol, L.G.T. Dados Multi e Hiperespectrais da Cultura da soja(Glycine max L.) e sua Relação com doses de gesso e Calcário no Solo. Master’s Thesis, Universidade Estadual de Maringá, Maringá, Brazil, 2017. [Google Scholar]
- Daughtry, C.S.T.; Gallo, K.P.; Goward, S.N.; Prince, S.D.; Kustas, W.P. Spectral estimates of absorbed radiation and phytomass production in corn and soybean canopies. Remote Sens. Environ. 1992, 39, 141–152. [Google Scholar] [CrossRef]
- Gitelson, A.A.; Peng, Y.; Arkebauer, T.J.; Suyker, A.E. Productivity, absorbed photosynthetically active radiation, and light use efficiency in crops: Implications for remote sensing of crop primary production. J. Plant Physiol. 2015, 177, 100–109. [Google Scholar] [CrossRef] [Green Version]
- Gitelson, A.A. Remote estimation of fraction of radiation absorbed by photosynthetically active vegetation: Generic algorithm for maize and soybean. Remote Sens. Lett. 2019, 10, 283–291. [Google Scholar] [CrossRef]
- Roujean, J.L.; Breon, F.M. Estimating PAR absorbed by vegetation from bidirectional reflectance measurements. Remote Sens. Environ. 1995, 51, 375–384. [Google Scholar] [CrossRef]
- Singer, J.W.; Meek, D.W.; Sauer, T.J.; Prueger, J.H.; Hatfield, J.L. Variability of light interception and radiation use efficiency in maize and soybean. Field Crops Res. 2011, 121, 147–152. [Google Scholar] [CrossRef]
- Damm, A.; Paul-Limoges, E.; Haghighi, E.; Simmer, C.; Morsdorf, F.; Schneider, F.D.; Tol, C.V.D.; Migliavacca, M.; Rascher, U. Remote sensing of plant-water relations: An overview and future perspectives. J. Plant Physiol. 2018, 227, 3–19. [Google Scholar] [CrossRef]
- Gates, D.M.; Keegan, H.J.; Schleter, J.C.; Weidner, V.R. Spectral properties of plants. Appl. Opt. 1965, 4, 11–20. [Google Scholar] [CrossRef]
- Latimer, P. Apparent shifts of absorption bands of cell suspensions and selective light scattering. Science 1958, 127, 29–30. [Google Scholar] [CrossRef] [PubMed]
- Ustin, S.L.; Jacquemoud, S.; Govaerts, Y. Simulation of photon transport in a three-dimensional leaf: Implications for photosynthesis. Plant Cell Environ. 2001, 24, 1095–1103. [Google Scholar] [CrossRef] [Green Version]
- Gao, B.C. NDWI—A normalized difference water index for remote sensing of vegetation liquid water from space. Remote Sens. Environ. 1996, 58, 257–266. [Google Scholar] [CrossRef]
- Wang, L.; Qu, J.J. NMDI: A normalized multi-band drought index for monitoring soil and vegetation moisture with satellite remote sensing. Geophys. Res. Lett. 2007, 34, L20405. [Google Scholar] [CrossRef]
- Zhang, Z.; Tang, B.H.; Li, Z.L. Retrieval of leaf water content from remotely sensed data using a vegetation index model constructed with shortwave infrared reflectances. Int. J. Remote Sens. 2019, 40, 2313–2323. [Google Scholar] [CrossRef]
- Wang, J.; Xu, R.; Yang, S. Estimation of plant water content by spectral absorption features centered at 1450 nm and 1940 nm regions. Environ. Monit. Assess. 2009, 157, 459. [Google Scholar] [CrossRef] [PubMed]
- Foster, A.J.; Kakani, V.G.; Ge, J.; Gregory, M.; Mosali, J. Discriminant analysis of nitrogen treatments in switchgrass and high biomass sorghum using leaf and canopy-scale reflectance spectroscopy. Int. J. Remote Sens. 2016, 37, 2252–2279. [Google Scholar] [CrossRef]
- Guzmán, Q.; Rivard, B.; Sánchez-Azofeifa, G.A. Discrimination of liana and tree leaves from a Neotropical Dry Forest using visible-near infrared and longwave infrared reflectance spectra. Remote Sens. Environ. 2018, 219, 135–144. [Google Scholar] [CrossRef]
- He, Y.; Li, X.; Deng, X. Discrimination of varieties of tea using near infrared spectroscopy by principal component analysis and BP model. J. Food Eng. 2007, 79, 1238–1242. [Google Scholar] [CrossRef]
- He, Y.; Li, X.; Shao, Y. Fast discrimination of apple varieties using Vis/NIR spectroscopy. Int. J. Food Prop. 2007, 10, 9–18. [Google Scholar] [CrossRef]
- Shirzadifar, A.; Bajwa, S.; Mireei, S.A.; Howatt, K.; Nowatzki, J. Weed species discrimination based on SIMCA analysis of plant canopy spectral data. Biosyst. Eng. 2018, 171, 143–154. [Google Scholar] [CrossRef]
- Holden, H.; LeDrew, E. Spectral discrimination of healthy and non-healthy corals based on cluster analysis, principal components analysis, and derivative spectroscopy. Remote Sens. Environ. 1998, 65, 217–224. [Google Scholar] [CrossRef]
- Lebow, P.K.; Brunner, C.C.; Maristany, A.G.; Butler, D.A. Classification of wood surface features by spectral reflectance. Wood and Fiber Sci. 2007, 28, 74–90. [Google Scholar]
- Clark, M.L.; Roberts, D.A.; Clark, D.B. Hyperspectral discrimination of tropical rain forest tree species at leaf to crown scales. Remote Sens. Environ. 2005, 96, 375–398. [Google Scholar] [CrossRef]
- Bravo, C.; Moshou, D.; West, J.; McCartney, A.; Ramon, H. Early disease detection in wheat fields using spectral reflectance. Biosyst. Eng. 2003, 84, 137–145. [Google Scholar] [CrossRef]
- Adam, E.; Mutanga, O. Spectral discrimination of papyrus vegetation (Cyperus papyrus L.) in swamp wetlands using field spectrometry. ISPRS J. Photogramm. Remote Sens. 2009, 64, 612–620. [Google Scholar] [CrossRef]
- Bajwa, S.; Rupe, J.; Mason, J. Soybean disease monitoring with leaf reflectance. Remote Sens. 2017, 9, 127. [Google Scholar] [CrossRef] [Green Version]
- Jin, X.L.; Wang, K.R.; Xiao, C.H.; Diao, W.Y.; Wang, F.Y.; Chen, B.; Li, S.K. Comparison of two methods for estimation of leaf total chlorophyll content using remote sensing in wheat. Field Crops Res. 2012, 135, 24–29. [Google Scholar] [CrossRef]
- Thenkabail, P.S.; Smith, R.B.; De Pauw, E. Hyperspectral vegetation indices and their relationships with agricultural crop characteristics. Remote Sens. Environ. 2000, 71, 158–182. [Google Scholar] [CrossRef]
- Fassnacht, F.E.; Latifi, H.; Stereńczak, K.; Modzelewska, A.; Lefsky, M.; Waser, L.T.; Straub, C.; Ghosh, A. Review of studies on tree species classification from remotely sensed data. Remote Sens. Environ. 2016, 186, 64–87. [Google Scholar] [CrossRef]
- Fehr, W.R.; Caviness, C.E. Stages of Soybean Development; Special Report 80; Iowa State University of Science and Technology: Ames, IA, USA, 1977. [Google Scholar]
- Honna, P.T.; Fuganti-Pagliarini, R.; Ferreira, L.C.; Molinari, M.D.; Marin, S.R.; de Oliveira, M.C.; Farias, J.R.B.; Neumaier, N.; Mertz-Henning, L.M.; Kanamori, N.; et al. Molecular, physiological, and agronomical characterization, in greenhouse and in field conditions, of soybean plants genetically modified with AtGolS2 gene for drought tolerance. Mol. Breed. 2016, 36, 157. [Google Scholar] [CrossRef] [Green Version]
- De Paiva Rolla, A.A.; Carvalho, J.D.F.C.; Fuganti-Pagliarini, R.; Engels, C.; Do Rio, A.; Marin, S.R.R.; de Oliveira, M.C.N.; Beneventi, M.A.; Marcelino-Guimarães, F.C.; Farias, J.R.B.; et al. Phenotyping soybean plants transformed with rd29A: AtDREB1A for drought tolerance in the greenhouse and field. Transgenic Res. 2014, 23, 75–87. [Google Scholar]
- Stolf-Moreira, R.; Lemos, E.G.M.; Carareto-Alves, L.; Marcondes, J.; Pereira, S.S.; Rolla, A.A.P.; Pereira, R.M.; Neumaier, N.; Binneck, E.; Abdelnoor, R.V.; et al. Transcriptional profiles of roots of different soybean genotypes subjected to drought stress. Plant Mol. Biol. Rep. 2011, 29, 19–34. [Google Scholar] [CrossRef]












| Spectral Dataset | Field Experimental Treatments | Days of Assessment | Genotype | Samples | Total Dataset Samples |
|---|---|---|---|---|---|
| Embrapa 2016/2017 | Irrigated; Non-irrigated; Drought stress in vegetative and reproductive stages | 7 | G1 | 112 | 444 |
| G2 | 108 | ||||
| G3 | 112 | ||||
| G4 | 112 | ||||
| - | - | ||||
| Embrapa 2017/2018 | Irrigated; Non-irrigated; Drought stress in vegetative and reproductive stages | 8 | G1 | 128 | 640 |
| G2 | 128 | ||||
| G3 | 128 | ||||
| G4 | 128 | ||||
| G5 | 128 | ||||
| Embrapa 2018/2019 [I] | Natural rainfall | 9 | G1 | 180 | 900 |
| G2 | 180 | ||||
| G3 | 180 | ||||
| G4 | 180 | ||||
| G5 | 180 | ||||
| Embrapa 2018/2019 [II] | Irrigated; Non-irrigated; Drought stress in vegetative and reproductive stages | 9 | G6 | 144 | 720 |
| G7 | 144 | ||||
| G8 | 144 | ||||
| G9 | 144 | ||||
| G10 | 144 | ||||
| Embrapa 2016–2019 | Irrigated; Non-irrigated; Drought stress in vegetative and reproductive stages | 24 | G1 | 420 | 1984 |
| G2 | 416 | ||||
| G3 | 420 | ||||
| G4 | 420 | ||||
| G5 | 308 | ||||
Unoeste 2018/2019 [I] | Natural rainfall | 4 | G1 | 105 | 525 |
| G2 | 105 | ||||
| G3 | 105 | ||||
| G4 | 105 | ||||
| G5 | 105 | ||||
| Unoeste 2018/2019 [II] | Natural rainfall | 4 | G6 | 105 | 420 |
| - | - | ||||
| G8 | 105 | ||||
| G9 | 105 | ||||
| G10 | 105 | ||||
| Embrapa 2016–2019 + Unoeste [I] | Irrigated; Non-irrigated; Drought stress in vegetative and reproductive stages | 28 | G1 | 525 | 2509 |
| G2 | 521 | ||||
| G3 | 525 | ||||
| G4 | 525 | ||||
| G5 | 413 | ||||
| Embrapa 2018/2019 + Unoeste [II] | Irrigated; Non-irrigated; Drought stress in vegetative and reproductive stages | 13 | G6 | 249 | 1140 |
| G7 | 144 | ||||
| G8 | 249 | ||||
| G9 | 249 | ||||
| G10 | 249 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Crusiol, .G.T.; Nanni, M.R.; Furlanetto, R.H.; Sibaldelli, R.N.R.; Cezar, E.; Sun, L.; Foloni, J.S.S.; Mertz-Henning, L.M.; Nepomuceno, A.L.; Neumaier, N.; et al. Classification of Soybean Genotypes Assessed Under Different Water Availability and at Different Phenological Stages Using Leaf-Based Hyperspectral Reflectance. Remote Sens. 2021, 13, 172. https://0-doi-org.brum.beds.ac.uk/10.3390/rs13020172
Crusiol GT, Nanni MR, Furlanetto RH, Sibaldelli RNR, Cezar E, Sun L, Foloni JSS, Mertz-Henning LM, Nepomuceno AL, Neumaier N, et al. Classification of Soybean Genotypes Assessed Under Different Water Availability and at Different Phenological Stages Using Leaf-Based Hyperspectral Reflectance. Remote Sensing. 2021; 13(2):172. https://0-doi-org.brum.beds.ac.uk/10.3390/rs13020172
Chicago/Turabian StyleCrusiol, Luis Guilherme Teixeira, Marcos Rafael Nanni, Renato Herrig Furlanetto, Rubson Natal Ribeiro Sibaldelli, Everson Cezar, Liang Sun, José Salvador Simonetto Foloni, Liliane Marcia Mertz-Henning, Alexandre Lima Nepomuceno, Norman Neumaier, and et al. 2021. "Classification of Soybean Genotypes Assessed Under Different Water Availability and at Different Phenological Stages Using Leaf-Based Hyperspectral Reflectance" Remote Sensing 13, no. 2: 172. https://0-doi-org.brum.beds.ac.uk/10.3390/rs13020172