The Horizontal Distribution of Branch Biomass in European Beech: A Model Based on Measurements and TLS Based Proxies
Abstract
:1. Introduction
2. Materials and Methods
2.1. Destructive Biomass Measurements
2.2. Modelling the Individual Horizontal Woody Branch Biomass Distribution
2.3. TLS Data Collection and Processing
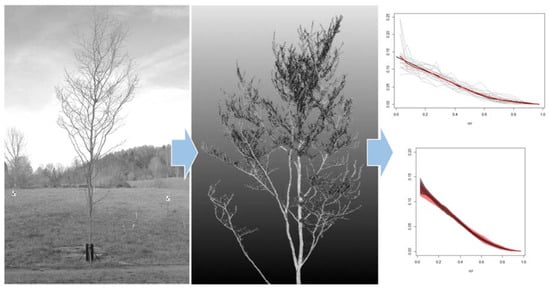
2.4. A New TLS Metric to Approximate Branch HBD
- The outer parts of the crown receive more hits than the inner parts.
- After having removed the returns from the stem, as described in Section 2.3, the first order branches do not, of course, emerge directly from the one-dimensional stem axis but at a certain distance—the stem radius—which becomes smaller towards the top of the tree, as shown in Figure 4.
- The outermost branches occlude the inner part of the crown.
- The stem axis does not follow a perfect upright straight line and it has not been perfectly vertically erected on the stem center on the ground.
- The horizontal distance to the stem axis (ρ) was calculated for each individual return by transforming the Cartesian coordinates of the original TLS point cloud to cylindrical coordinates (ρ, φ, z) with φ = azimuth and z = height:
- The crown returns were classified according to the four quadrants defined by the X and Y Cartesian coordinates in the horizontal plane (see Figure 5), and separate histograms were produced in each quadrant in 1 cm steps of ρ.
- From steps one and two, the distribution of TLS returns can be determined for each quadrant. The starting point for each quadrant (ρSTART) was set to half of the range in ρ between 0 and the center of the bin with maximum observed frequency (Figure 6A,B). The entire histogram was then moved to the left by setting ρSTART as the starting point of the histogram (Figure 6C). The order for the k bins at the left of ρSTART was inverted, and the counts summed to the first k remaining bins (Figure 6D,E) so that the total number of hits captured in the distribution remained the same.
- The resulting histograms for each quadrant (Figure 6E) were combined into a single histogram and then standardized in ρ relative to the maximum ρ across all quadrants. This is what we finally refer to as the standardized composite histogram (SCH).
- The SCH was then grouped into new classes of relative ρ, and the counts were standardized with the total number of counts over all ρ classes.
2.5. Evaluation of the TLS Metric SCH
3. Results
3.1. Empirical Branch HBD
3.2. The SCH TLS Metric as a Proxy for the Branch HBD
4. Discussion
5. Conclusions
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ketterings, Q.M.; Coe, R.; van Noordwijk, M.; Ambagau, Y.; Palm, C.A. Reducing Uncertainty in the use of Allometric Biomass Equations for Predicting Above-Ground Tree Biomass in Mixed Secondary Forests. For. Ecol. Manag. 2001, 146, 199–209. [Google Scholar] [CrossRef]
- Pérez-Cruzado, C.; Rodríguez Soalleiro, R. Improvement in Accuracy of Aboveground Biomass Estimation in Eucalyptus Nitens Plantations: Effect of Bole Sampling Intensity and Explanatory Variables. For. Ecol. Manag. 2011, 261, 2016–2028. [Google Scholar] [CrossRef]
- Zianis, D.; Muukkonen, P.; Mäkipää, R.; Mencuccini, M. Biomass and Stem Volume Equations for Tree Species in Europe; Tammer-Paino Oy: Tampere, Finland, 2005; p. 63. [Google Scholar]
- Ruiz-González, A.D.; Álvarez-González, J.G. Canopy Bulk Density and Canopy Base Height Equations for Assessing Crown Fire Hazard in Pinus Radiata Plantations. Can. J. For. Res. 2011, 41, 839–850. [Google Scholar] [CrossRef]
- Jiménez, E.; Vega, J.A.; Ruiz-González, A.D.; Guijarro, M.; Alvarez-González, J.G.; Madrigal, J.; Cuiñas, P.; Hernando, C.; Fernández-Alonso, J.M. Carbon Emissions and Vertical Pattern of Canopy Fuel Consumption in Three Pinus pinaster Ait. Active Crown Fires in Galicia (NW Spain). Ecol. Eng. 2013, 54, 202–209. [Google Scholar] [CrossRef]
- Affleck, D.L.R.; Keyes, C.R.; Goodburn, J.M. Conifer Crown Fuel Modeling: Current Limits and Potential for Improvement. West. J. Appl. For. 2012, 27, 165–169. [Google Scholar] [CrossRef] [Green Version]
- Kleinn, C.; Magnussen, S.; Nölke, N.; Magdon, P.; Álvarez-González, J.G.; Fehrmann, L.; Pérez-Cruzado, C. Improving Precision of Field Inventory Estimation of Aboveground Biomass through an Alternative View on Plot Biomass. For. Ecosyst. 2020, 7, 1–10. [Google Scholar] [CrossRef]
- Næsset, E. Predicting Forest Stand Characteristics with Airborne Scanning Laser using a Practical Two-Stage Procedure and Field Data. Remote Sens. Environ. 2002, 80, 88–99. [Google Scholar] [CrossRef]
- Tahvanainen, T.; Forss, E. Individual Tree Models for the Crown Biomass Distribution of Scots Pine, Norway Spruce and Birch in Finland. For. Ecol. Manag. 2008, 255, 455–467. [Google Scholar] [CrossRef]
- Nemec, A.F.L.; Parish, R.; Goudie, J.W. Modelling Number, Vertical Distribution, and Size of Live Branches on Coniferous Tree Species in British Columbia. Can. J. For. Res. 2012, 42, 1072–1090. [Google Scholar] [CrossRef]
- Kershaw, J.A.; Maguire, D.A. Crown Structure in Western Hemlock, Douglas-Fir, and Grand Fir in Western Washington: Trends in Branch-Level Mass and Leaf Area. Can. J. For. Res. 1996, 25, 1897–1912. [Google Scholar] [CrossRef]
- Xu, M.; Harrington, T.B. Foliage Biomass Distribution of Loblolly Pine as Affected by Tree Dominance, Crown Size, and Stand Characteristics. Can. J. For. Res. 1998, 28, 887–892. [Google Scholar] [CrossRef]
- Mascaro, J.; Detto, M.; Asner, G.P.; Muller-Landau, H.C. Evaluating Uncertainty in Mapping Forest Carbon with Airborne LiDAR. Remote Sens. Environ. 2011, 115, 3770–3774. [Google Scholar] [CrossRef]
- Nielsen, C.C.N.; Mackenthum, G. Die horizontale Varia-tion der Feinwurzelintensität in Waldböden in Abhängigkeit vonder Bestockungsdichte. Einerechnerische Methode zur Bestimmung der “Wurzelintensitätsglocke” an Einzelbäumen. Allg. Forst. Jagdztg. 1991, 162, 112–119. [Google Scholar]
- Fehrmann, L.; Kuhr, M.; von Gadow, K. Zur Analyse Der Grobwurzelsysteme Großer Waldbäume and Fichte [Picea abies (L.) Karst.] Und Buche [Fagus sylvatica L.] (In German: “Analyis of the Coarse Root Systems of Large Trees at Spruce [Picea abies (L.) Karst.] and Beech [Fagus sylvatica L.]”). Forstarchiv 2003, 74, 96–102. [Google Scholar]
- Liang, X.; Kankare, V.; Hyyppä, J.; Wang, Y.; Kukko, A.; Haggrén, H.; Yu, X.; Kaartinen, H.; Jaakkola, A.; Guan, F. Terrestrial Laser Scanning in Forest Inventories. ISPRS J. Photogramm. Remote Sens. 2016, 115, 63–77. [Google Scholar] [CrossRef]
- Zhang, W.; Wan, P.; Wang, T.; Cai, S.; Chen, Y.; Jin, X.; Yan, G. A Novel Approach for the Detection of Standing Tree Stems from Plot-Level Terrestrial Laser Scanning Data. Remote Sens. 2019, 11, 211. [Google Scholar] [CrossRef] [Green Version]
- Pitkänen, T.P.; Raumonen, P.; Kangas, A. Measuring Stem Diameters with TLS in Boreal Forests by Complementary Fitting Procedure. ISPRS J. Photogramm. Remote Sens. 2019, 147, 294–306. [Google Scholar] [CrossRef]
- Zhu, Z.; Kleinn, C.; Nölke, N. Towards Tree Green Crown Volume: A Methodological Approach using Terrestrial Laser Scanning. Remote Sens. 2020, 12, 1841. [Google Scholar] [CrossRef]
- LaRue, E.A.; Wagner, F.W.; Fei, S.; Atkins, J.W.; Fahey, R.T.; Gough, C.M.; Hardiman, B.S. Compatibility of Aerial and Terrestrial LiDAR for Quantifying Forest Structural Diversity. Remote Sens. 2020, 12, 1407. [Google Scholar] [CrossRef]
- Guan, H.; Su, Y.; Sun, X.; Xu, G.; Li, W.; Ma, Q.; Wu, X.; Wu, J.; Liu, L.; Guo, Q. A Marker-Free Method for Registering Multi-Scan Terrestrial Laser Scanning Data in Forest Environments. ISPRS J. Photogramm. Remote Sens. 2020, 166, 82–94. [Google Scholar] [CrossRef]
- Calders, K.; Adams, J.; Armston, J.; Bartholomeus, H.; Bauwens, S.; Bentley, L.P.; Chave, J.; Danson, F.M.; Demol, M.; Disney, M.; et al. Terrestrial Laser Scanning in Forest Ecology: Expanding the Horizon. Remote Sens. Environ. 2020, 251, 112102. [Google Scholar] [CrossRef]
- Hauglin, M.; Astrup, R.; Gobakken, T.; Næsset, E. Estimating Single-Tree Branch Biomass of Norway Spruce with Terrestrial Laser Scanning using Voxel-Based and Crown Dimension Features. Scand. J. For. Res. 2013, 28, 456–469. [Google Scholar] [CrossRef]
- Cifuentes, R.; Van der Zande, D.; Farifteh, J.; Salas, C.; Coppin, P. Effects of Voxel Size and Sampling Setup on the Estimation of Forest Canopy Gap Fraction from Terrestrial Laser Scanning Data. Agric. For. Meteorol. 2014, 194, 230–240. [Google Scholar] [CrossRef]
- Grau, E.; Durrieu, S.; Fournier, R.; Gastellu-Etchegorry, J.P.; Yin, T. Estimation of 3D Vegetation Density with Terrestrial Laser Scanning Data using Voxels. A Sensitivity Analysis of Influencing Parameters. Remote Sens. Environ. 2017, 191, 373–388. [Google Scholar] [CrossRef]
- Hackenberg, J.; Spiecker, H.; Calders, K.; Disney, M.; Raumonen, P. SimpleTree—An Efficient Open Source Tool to Build Tree Models from TLS Clouds. Forests 2015, 6, 4245–4294. [Google Scholar] [CrossRef]
- Lau, A.; Bentley, L.P.; Martius, C.; Shenkin, A.; Bartholomeus, H.; Raumonen, P.; Malhi, Y.; Jackson, T.; Herold, M. Quantifying Branch Architecture of Tropical Trees using Terrestrial LiDAR and 3D Modelling. Trees 2018, 32, 1219–1231. [Google Scholar] [CrossRef] [Green Version]
- Park, H.; Lim, S.; Trinder, J.; Turner, R. 3D Surface Reconstruction of Terrestrial Laser Scanner Data for Forestry. In Proceedings of the 2010 IEEE International Geoscience and Remote Sensing Symposium, Honolulu, HI, USA, 25–30 July 2010; pp. 4366–4369. [Google Scholar]
- Raumonen, P.; Kaasalainen, M.; Åkerblom, M.; Kaasalainen, S.; Kaartinen, H.; Vastaranta, M.; Holopainen, M.; Disney, M.; Lewis, P. Fast Automatic Precision Tree Models from Terrestrial Laser Scanner Data. Remote Sens. 2013, 5, 491–520. [Google Scholar] [CrossRef] [Green Version]
- Disney, M.I.; Boni Vicari, M.; Burt, A.; Calders, K.; Lewis, S.L.; Raumonen, P.; Wilkes, P. Weighing Trees with Lasers: Advances, Challenges and Opportunities. Interface Focus 2018, 8, 20170048. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Calders, K.; Newnham, G.; Burt, A.; Murphy, S.; Raumonen, P.; Herold, M.; Culvenor, D.; Avitabile, V.; Disney, M.; Armston, J.; et al. Nondestructive Estimates of Above-ground Biomass using Terrestrial Laser Scanning. Methods Ecol. Evol. 2015, 6, 198–208. [Google Scholar] [CrossRef]
- Hackenberg, J.; Wassenberg, M.; Spiecker, H.; Sun, D. Non Destructive Method for Biomass Prediction Combining TLS Derived Tree Volume and Wood Density. Forests 2015, 6, 1274–1300. [Google Scholar] [CrossRef]
- Dassot, M.; Colin, A.; Santenoise, P.; Fournier, M.; Constant, T. Terrestrial Laser Scanning for Measuring the Solid Wood Volume, Including Branches, of Adult Standing Trees in the Forest Environment. Comput. Electron. Agric. 2012, 89, 86–93. [Google Scholar] [CrossRef]
- Abegg, M.; Kükenbrink, D.; Zell, J.; Schaepman, M.E.; Morsdorf, F. Terrestrial Laser Scanning for Forest Inventories—Tree Diameter Distribution and Scanner Location Impact on Occlusion. Forests 2017, 8, 184. [Google Scholar] [CrossRef] [Green Version]
- Max, T.A.; Burkhart, H.E. Segmented Polynomial Regression Applied to Taper Equations. For. Sci. 1976, 22, 283–289. [Google Scholar]
- Van Laar, A.; Akça, A. Forest Mensuration; Springer Science & Business Media: Berlin/Heidelberg, Germany, 2007. [Google Scholar]
- Burkhart, H.E.; Tomé, M. Modeling Forest Trees and Stands; Springer Science & Business Media: Dordercht, The Netherlands, 2012. [Google Scholar]
- Weiskittel, A.; Li, R. Development of Regional Taper and Volume Equations: Hardwood Species; Roth, B.E., Ed.; Cooperative Forestry Research Unit: 2011 Annual Report; University of Maine: Orono, ME, USA, 2012; pp. 76–84. [Google Scholar]
- Pérez-Cruzado, C.; Fehrmann, L.; Magdon, P.; Cañellas, I.; Sixto, H.; Kleinn, C. On the Site-Level Suitability of Biomass Models. Environ. Model. Softw. 2015, 73, 14–26. [Google Scholar] [CrossRef]
- Durbin, J.; Watson, G.S. Testing for Serial Correlation in Least Squares Regression: I. Biometrika 1950, 37, 409–428. [Google Scholar] [PubMed]
- Durbin, J.; Watson, G.S. Testing for Serial Correlation in Least Squares Regression. II. Biometrika 1951, 38, 159–177. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2018. [Google Scholar]
- Enquist, B.J. Universal Scaling in Tree and Vascular Plant Allometry: Toward a General Quantitative Theory Linking Plant Form and Function from Cells to Ecosystems. Tree Physiol. 2002, 22, 1045–1064. [Google Scholar] [CrossRef] [Green Version]
- Demaerschalk, J.P. Converting Volume Equations to Compatible Taper Equations. For. Sci. 1972, 18, 241–245. [Google Scholar] [CrossRef]
- Fang, Z.; Bailey, R.L. Compatible Volume and Taper Models with Coefficients for Tropical Species on Hainan Island in Southern China. For. Sci. 1999, 45, 85–100. [Google Scholar]
- Muhairwe, C.K. Taper Equations for Eucalyptus Pilularis and Eucalyptus Grandis for the North Coast in New South Wales, Australia. For. Ecol. Manag. 1999, 113, 251–269. [Google Scholar] [CrossRef]
- Fang, Z.; Borders, B.E.; Bailey, R.L. Compatible Volume-Taper Models for Lobolly and Slash Pine Based on a System with Segmented-Stem Form Factors. For. Sci. 2000, 46, 1–12. [Google Scholar]
- Castedo Dorado, F.; Dieguez-Aranda, U.; Barrio Anta, M.; Sánchez Rodríguez, M.; von Gadow, K. A Generalized Height-Diameter Model Including Random Components for Radiata Pine Plantations in Northwestern Spain. For. Ecol. Manag. 2006, 229, 202–213. [Google Scholar] [CrossRef]
- Westfall, J.A.; Scott, C.T. Taper Models for Commercial Tree Species in the Northeastern United States. For. Sci. 2010, 56, 515–528. [Google Scholar]
- Tang, X.; Pérez-Cruzado, C.; Fehrmann, L.; Álvarez-González, J.G.; Lu, Y.; Kleinn, C. Development of a Compatible Taper Function and Stand-Level Merchantable Volume Model for Chinese Fir Plantations. PLoS ONE 2016, 11, e0147610. [Google Scholar] [CrossRef] [PubMed] [Green Version]










| dbh (cm) | h (m) | Crown Ratio CR | Crown Diameter CD (m) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Range | Mean | SD | Range | Mean | SD | Range | Mean | SD | Range | Mean | SD | |
| (1) Whole stand | 5.0–23.9 | 10.3 | 3.7 | 6.3–21.1 | 11.6 | 2.0 | - | - | - | - | - | - |
| (2) Empirical data | 9.2–20.6 | 14.0 | 3.0 | 13.3–21.1 | 17.7 | 2.2 | 0.437–0.814 | 0.612 | 0.082 | 1.84–9.28 | 3.73 | 1.65 |
| (3) TLS | 9.2–20.6 | 14.3 | 3.1 | 13.9–21.1 | 17.8 | 2.2 | 0.437–0.814 | 0.617 | 0.087 | 1.84–9.28 | 3.85 | 1.82 |
| Model (n = 460) | Parameter | Estimate | Std. Error |
|---|---|---|---|
| Global model (Equation (1)) | b1 | −0.0047 | 0.0014 |
| b2 | 0.0024 | 0.0007 | |
| b3 | 0.0366 | 0.0026 | |
| a | 0.6654 | 0.0208 | |
| rho | 0.3584 | 0.0436 | |
| Generalized model (Equation (2)) | b1 | −0.0053 | 0.0014 |
| b2 | 0.0027 | 0.0007 | |
| b31 | 0.0679 | 0.0098 | |
| b32 | −0.0497 | 0.0142 | |
| a | 0.6541 | 0.0213 | |
| rho | 0.3429 | 0.0440 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pérez-Cruzado, C.; Kleinn, C.; Magdon, P.; Álvarez-González, J.G.; Magnussen, S.; Fehrmann, L.; Nölke, N. The Horizontal Distribution of Branch Biomass in European Beech: A Model Based on Measurements and TLS Based Proxies. Remote Sens. 2021, 13, 1041. https://0-doi-org.brum.beds.ac.uk/10.3390/rs13051041
Pérez-Cruzado C, Kleinn C, Magdon P, Álvarez-González JG, Magnussen S, Fehrmann L, Nölke N. The Horizontal Distribution of Branch Biomass in European Beech: A Model Based on Measurements and TLS Based Proxies. Remote Sensing. 2021; 13(5):1041. https://0-doi-org.brum.beds.ac.uk/10.3390/rs13051041
Chicago/Turabian StylePérez-Cruzado, César, Christoph Kleinn, Paul Magdon, Juan Gabriel Álvarez-González, Steen Magnussen, Lutz Fehrmann, and Nils Nölke. 2021. "The Horizontal Distribution of Branch Biomass in European Beech: A Model Based on Measurements and TLS Based Proxies" Remote Sensing 13, no. 5: 1041. https://0-doi-org.brum.beds.ac.uk/10.3390/rs13051041