Examining the Spectral Separability of Prosopis glandulosa from Co-Existent Species Using Field Spectral Measurement and Guided Regularized Random Forest
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Area

2.2. Identification of Mesquite and Other Co-Existent Tree Species
2.3. Field Spectroscopy Measurements

| Species | Training Samples (70%) | Test Samples (30%) | Total Samples |
|---|---|---|---|
| Prosopis glandulosa (PR) | 93 | 40 | 133 |
| Acacia karroo (AK) | 76 | 32 | 108 |
| Acacia mellifera (AM) | 93 | 40 | 133 |
| Ziziphus mucronata (ZM) | 87 | 37 | 124 |
2.4. Field Spectroscopy Data Analysis

2.5. Random Forest Classifier and Variable Importance Measurement
2.6. Feature Selection Using Guided Regularized Random Forest
2.7. Accuracy Assessment
3. Results
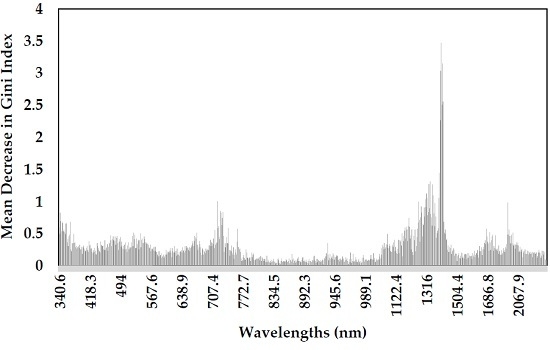
3.1. Variables Importance Measurement and Selection


3.2. Accuracy Assessment
| Class | Using 1825 Wavelengths | Class | Using the Selected 11 Wavelengths | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| AK | AM | PR | ZM | Total | AK | AM | PR | ZM | Total | ||
| AK | 25 | 2 | 3 | 2 | 32 | AK | 27 | 1 | 2 | 2 | 37 |
| AM | 2 | 34 | 3 | 1 | 40 | AM | 2 | 36 | 2 | 0 | 40 |
| PR | 4 | 3 | 30 | 3 | 40 | PR | 1 | 1 | 36 | 2 | 40 |
| ZM | 3 | 1 | 4 | 29 | 37 | ZM | 2 | 0 | 2 | 33 | 37 |
| Total | 34 | 40 | 40 | 35 | 149 | Total | 32 | 38 | 42 | 37 | 149 |
| OA = 79.19% | OA = 88.59% | ||||||||||
| Kappa = 0.7201 | Kappa = 0.8524 | ||||||||||
| Class | Using 1825 Wavelengths | Class | Using 11 Wavelengths | ||
|---|---|---|---|---|---|
| Producer’s Accuracy (%) | User’s Accuracy (%) | Producer’s Accuracy (%) | User’s Accuracy (%) | ||
| AK | 73.53 | 78.13 | AK | 84.38 | 84.38 |
| AM | 85.00 | 85.00 | AM | 94.74 | 90.00 |
| PR | 75.00 | 75.00 | PR | 85.71 | 90.00 |
| ZM | 82.86 | 78.38 | ZM | 89.19 | 89.19 |
4. Discussion
5. Conclusions
- One of the major problems in controlling mesquite has been the presence of mixed stands that consist of alien Prosopis mixed and indigenous species. Prosopis glandulosa can be accurately detected from its co-existent species, namely Acacia karroo, Acacia mellifera and Ziziphus mucronata, using hyperspectral data. Such potential data could provide environmental managers and ecologists insight into the development of possible appropriate spatio-temporal management practices to better control the invasive spread of mesquite.
- The problem of high dimensionality associated with spectroscopy data processing can be reduced considerably by making use of the new-developed GRRF method. The new GRRF method created high quality feature variables for the traditional RF classifier and can thus be seen as a more efficient and effective feature selection tool to reduce the high dimensionality in spectroscopy data. However, this assertion should receive considerable additional testing and comparison with the commonly-used variable selection methods before it is accepted as a substitute for reliable high dimensionality reduction.
- The wavelengths selected by GRRF showed the greatest discriminatory power of Prosopis from other species across the spectrum regions, mainly visible, red edge and short-wave infrared regions. These wavelengths are located at 356.3 nm, 468.5 nm, 531.1 nm, 665.2 nm, 1262.3 nm, 1354.1 nm, 1361.7 nm, 1376.9 nm, 1407.1 nm, 1410.9 nm and 1414.6 nm.
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Shackleton, R.T.; Le Maitre, D.C.; Pasiecznik, N.M.; Richardson, D.M. Prosopis: A global assessment of the biogeography, benefits, impacts and management of one of the world's worst woody invasive plant taxa. AoB Plants 2014, 6. [Google Scholar] [CrossRef] [PubMed]
- Zeila, A. Mapping and managing the spread of prosopis juliflora in Garissa County, Kenya. Ph.D. Thesis, Kenyatta University, Nairobi, Kenya, 2011. [Google Scholar]
- Klinken, R.; Shepherd, D.; Parr, R.; Robinson, T.; Anderson, L. Mapping mesquite (prosopis) distribution and density using visual aerial surveys. Rangel. Ecol. Manag. 2007, 60, 408–416. [Google Scholar] [CrossRef]
- Awale, A.I.; Sugule, A.J. Proliferation of Honey Mesquite (Prosopis juliflora) in Somaliland: Opportunities and Challenges. Available online: http://agris.fao.org/agris-search/search.do?recordID=SO2009100043 (accessed on 14 February 2016).
- Lloyd, J.; Berg, E.; Badenhorst, N. Mapping the Spatial Distribution and Biomass of Prosopis in the Northern Cape Province, South Africa, with the Aid of Remote Sensing and Geographic Information Systems; Report No. GW/A/98/68; Agricultural Research Council-Institute for Soil, Climate and Water: Pretoria, South Africa, 2002. [Google Scholar]
- Pasiecznik, N.M.; Felker, P.; Association, H.D.R. The'prosopis Juliflora'-'Prosopis Pallida'complex: A Monograph; HDRA: Coventry, UK, 2001. [Google Scholar]
- Geesingis, D.; Al Khawlani, M.; Abba, M.L. Management of introduced prosopisspecies: Can economic exploitation control an invasive species? Unasylva 2004, 217, 36–44. [Google Scholar]
- Zimmermann, H. Biological control of mesquite, prosopis spp.(fabaceae), in South Africa. Agric. Ecosyst. Environ. 1991, 37, 175–186. [Google Scholar] [CrossRef]
- Ghazanfar, S. Invasive prosopis in Sultanate of Oman. Aliens 1996, 3, 10–11. [Google Scholar]
- Chikuni, M.; Dudley, C.; Sambo, E. Prosopis glandulosa torrey (leguminosae-mimosoidae) at Swang'oma, Lake Chilwa Plain: A blessing in disguise? Malawi J. Sci. Technol. 2005, 7, 10–16. [Google Scholar]
- Choge, S.; Clement, N.; Gitonga, M.; Okuye, J. Status Report on Commercialization of Prosopis Tree Resources in Kenya; KEFRI: Nairobi, Kenya, 2012. [Google Scholar]
- Laxén, J.P. Is Prosopis A Curse or A Blessing?: An Ecological Economic Analysis of An Invasive Alien Tree Species in Sudan; Tropical Forestry Reports; University of Helsinki: Helsinki, Finland, 2007. [Google Scholar]
- Elfadl, M.A.; Luukkanen, O. Field studies on the ecological strategies of Prosopis juliflora in a dryland ecosystem: A leaf gas exchange approach. J. Arid Environ. 2006, 66, 1–15. [Google Scholar] [CrossRef]
- Pasiecznik, N. Prosopis-pest or providence, weed or wonder tree? ETFRN 1999, 28, 14–15. [Google Scholar]
- Versfeld, D.; Le Maitre, D.; Chapman, R. Alien Invading Plants and Water Resources in South Africa: A Preliminary Assessment; WRC report (no. TT 99/98); Water Research Commission: Pretoria, South Africa, 1998. [Google Scholar]
- Berg, E.C.; Kotze, I.; Beukes, H. Detection, quantification and monitoring of prosopis in the Northern Cape Province of South Africa using remote sensing and GIS. South Afr. J. Geomat. 2014, 2, 68–81. [Google Scholar]
- Le Maitre, D.C.; Richardson, D.M.; Chapman, R.A. Alien plant invasions in south africa: Driving forces and the human dimension: Working for water. South Afr. J.Sci. 2004, 100, 103–112. [Google Scholar]
- Osmond, R. Mesquite Best Practice Manual: Control and Management Options for Mesquite (prosopis spp.) in Australia; Department of Natural Resources and Mines: Brisbane, Australia, 2003. [Google Scholar]
- Maundu, P.; Kibet, S.; Morimoto, Y.; Imbumi, M.; Adeka, R. Impact of prosopis juliflora on kenya's semi-arid and arid ecosystems and local livelihoods. Biodiversity 2009, 10, 33–50. [Google Scholar] [CrossRef]
- Witt, A. Impacts of invasive plants and their sustainable management in agro-ecosystems in Africa: A review. CABI Afr. 2010, 2010, 1102–1109. [Google Scholar]
- Mwangi, E.; Swallow, B. Invasion of Prosopis Juliflora and Local Livelihoods: Case Study from the Lake Baringo Area of Kenya; World Agroforestry Centre: Nairobi, Kenya, 2005. [Google Scholar]
- Baillie, J.; Hilton-Taylor, C.; Stuart, S.N. 2004 Iucn Red List of Threatened Species: A Global Species Assessment; IUCN: Gland, Switzerland, 2004. [Google Scholar]
- Harding, G. Status of prosopis as a weed. Appl. Plant Sci. Toegep. Plantwet. 1987, 1, 43–48. [Google Scholar]
- Coetzer, W.; Hoffmann, J. Establishment ofneltumius arizonensis (coleoptera: Bruchidae) on mesquite (prosopisspecies: Mimosaceae) in South Africa. Biol. Control 1997, 10, 187–192. [Google Scholar] [CrossRef]
- Zachariades, C.; Hoffmann, J.; Roberts, A. Biological control of mesquite (prosopis species)(fabaceae) in South Africa. Afr. Entomol. 2011, 19, 402–415. [Google Scholar] [CrossRef]
- Mampholo, R.K. To determine the extent of bush encroachment with focus on prosopis species on selected farms in the Vryburg district of north west province/by ramakgwale Klaas mampholo. Ph.D. Thesis, North-West University, Mahikeng, South Africa, 2006. [Google Scholar]
- Nie, W.; Yuan, Y.; Kepner, W.; Erickson, C.; Jackson, M. Hydrological impacts of mesquite encroachment in the upper San Pedro watershed. J. Arid Environ. 2012, 82, 147–155. [Google Scholar] [CrossRef]
- Hoshino, B.; Karamalla, A.; Manayeva, K.; Yoda, K.; Suliman, M.; Elgamri, M.; Nawata, H.; Yasuda, H. Evaluating the invasion strategic of mesquite (prosopis juliflora) in eastern Sudan using remotely sensed technique. J. Arid Land Stud. 2012, 22, 1–4. [Google Scholar]
- Barry, P.; Mendenhall, J.; Jarecke, P.; Folkman, M.; Pearlman, J.; Markham, B. EO-1 hyperion hyperspectral aggregation and comparison with EO-1 advanced land imager and landsat 7 ETM+. Proc. IEEE 2002. [Google Scholar] [CrossRef]
- Liew, S.C.; Chang, C.W.; Lim, K.H. Hyperspectral land cover classification of EO-1 hyperion data by principal component analysis and pixel unmixing. Proc. IEEE 2002. [Google Scholar] [CrossRef]
- Adam, E.; Mutanga, O.; Rugege, D.; Ismail, R. Discriminating the papyrus vegetation (cyperus papyrus l.) and its co-existent species using random forest and hyperspectral data resampled to hymap. Int. J. Remote Sens. 2012, 33, 552–569. [Google Scholar] [CrossRef]
- Artigas, F.; Yang, J. Hyperspectral remote sensing of marsh species and plant vigour gradient in the New Jersey Meadowlands. Int. J. Remote Sens. 2005, 26, 5209–5220. [Google Scholar] [CrossRef]
- Cho, M.A.; Sobhan, I.; Skidmore, A.K.; de Leeuw, J. Discriminating species using hyperspectral indices at leaf and canopy scales. Int. Arch. Spat. Infor. 2008, 2008, 369–376. [Google Scholar]
- Fung, T.; Ma, F.Y.; Sui, W.L. Hyperspectral data analysis for subtropical tree species identification. In Proceedings of the 1999 ASPRS Annual Conference, Portland, OR, USA, 2–6 June 1999.
- Kumar, L.; Skidmore, A.K. Use of derivative spectroscopy to identify regions of difference between some australian eucalypt species. In Proceedings of 9th Australasian Remote Sensing and Photogrammetry Conference, Sydney, Australia, 20–24 July 1998.
- Hsu, P. Feature extraction of hyperspectral images using wavelet and matching pursuit. ISPRS J. Photogramm. Remote Sens. 2007, 62, 78–92. [Google Scholar] [CrossRef]
- Melgani, F.; Bruzzone, L. Classification of hyperspectral remote sensing images with support vector machines. IEEE Trans. Geosci. Remote Sens. Lett. 2004, 42, 1778–1790. [Google Scholar] [CrossRef]
- Kavzoglu, T.; Mather, P.M. The role of feature selection in artificial neural network applications. Int. J. Remote Sens. 2002, 23, 2919–2937. [Google Scholar] [CrossRef]
- Bajcsy, P.; Groves, P. Methodology for hyperspectral band selection. Photogramm. Eng. Remote Sens. 2004, 70, 793–802. [Google Scholar] [CrossRef]
- Shaw, G.; Manolakis, D. Signal processing for hyperspectral image exploitation. IEEE Signal Proc. Mag. 2002, 19, 12–16. [Google Scholar] [CrossRef]
- Pal, M. Random forest classifier for remote sensing classification. Int. J. Remote Sens. 2005, 26, 217–222. [Google Scholar] [CrossRef]
- Borges, J.S.; Marcal, A.R.S.; Dias, J.M.B. Evaluation of feature extraction and reduction methods for hyperspectral images. In Proceedings of New Developments and Challenges in Remote Sensing, Rotterdam, the Netherlands, 29 May–2 June 2006.
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Strobl, C.; Boulesteix, A.; Kneib, T.; Augustin, T.; Zeileis, A. Conditional variable importance for random forests. BMC Bioinform. 2008, 9, 307. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deng, H.; Runger, G. Feature selection via regularized trees. 2012. [CrossRef]
- Guided Random Forest in the RRF Package. Available online: http://arxiv.org/abs/1306.0237 (accessed on 14 February 2016).
- Deng, H.; Runger, G. Gene selection with guided regularized random forest. Pattern Recognit. 2013, 46, 3483–3489. [Google Scholar] [CrossRef]
- Mucina, L.; Rutherford, M.C. The Vegetation of South Africa, Lesotho and Swaziland; South African National Biodiversity Institute: Strelitzia, South African, 2006. [Google Scholar]
- Adam, E.M.; Mutanga, O.; Rugege, D.; Ismail, R. Field spectrometry of papyrus vegetation (cyperus papyrus l.) in swamp wetlands of St Lucia, South Africa. Proc. IEEE 2009. [Google Scholar] [CrossRef]
- Hagos, M.; Smit, G. Soil enrichment by acacia mellifera subsp. Detinens on nutrient poor sandy soil in a semi-arid Southern African savanna. J. Arid Environ. 2005, 61, 47–59. [Google Scholar] [CrossRef]
- Smit, G.; Richter, C.; Aucamp, A. Bush encroachment: An approach to understanding and managing the problem. In Veld Management in South Africa; University of Natal Press: Scottsville, KY, USA, 1999; pp. 246–260. [Google Scholar]
- Taylor, C.; Barker, N. Species limits in vachellia (acacia) karroo (mimosoideae: Leguminoseae): Evidence from automated issr DNA “fingerprinting”. South Afr. J. Bot. 2012, 83, 36–43. [Google Scholar] [CrossRef]
- Priyanka, C.; Kumar, P.; Bankar, S.P.; Karthik, L. In vitro antibacterial activity and gas chromatography—Mass spectroscopy analysis of acacia karoo and ziziphus mauritiana extracts. J. Taibah Univ. Sci. 2015, 9, 13–19. [Google Scholar] [CrossRef]
- Spectral Evolution Rs-3500_User's Manual. Available online: http://www.spectralevolution.com/applications_Validate.html (accessed on 14 February 2016).
- Bian, M.; Skidmore, A.K.; Schlerf, M.; Wang, T.; Liu, Y.; Zeng, R.; Fei, T. Predicting foliar biochemistry of tea (camellia sinensis) using reflectance spectra measured at powder, leaf and canopy levels. ISPRS J. Photogramm. Remote Sens. 2013, 78, 148–156. [Google Scholar] [CrossRef]
- Milton, E.; Schaepman, M.; Anderson, K.; Kneubühler, M.; Fox, N. Progress in field spectroscopy. Remote Sens. Environ. 2009, 113, S92–S109. [Google Scholar] [CrossRef]
- Zhao, D.; Huang, L.; Li, J.; Qi, J. A comparative analysis of broadband and narrowband derived vegetation indices in predicting lai and ccd of a cotton canopy. ISPRS J. Photogramm. Remote Sens. 2007, 62, 25–33. [Google Scholar] [CrossRef]
- Lin, X.; Sun, L.; Li, Y.; Guo, Z.; Li, Y.; Zhong, K.; Wang, Q.; Lu, X.; Yang, Y.; Xu, G. A random forest of combined features in the classification of cut tobacco based on gas chromatography fingerprinting. Talanta 2011, 82, 1571–1575. [Google Scholar] [CrossRef] [PubMed]
- Genuer, R.; Poggi, J.-M.; Tuleau-Malot, C. Variable selection using random forests. Pattern Recognit. Lett. 2010, 31, 2225–2236. [Google Scholar] [CrossRef]
- Tian, F.; Yang, L.; Lv, F.; Zhou, P. Predicting liquid chromatographic retention times of peptides from the drosophila melanogaster proteome by machine learning approaches. Anal. Chimic. Acta 2009, 644, 10–16. [Google Scholar] [CrossRef] [PubMed]
- Strobl, C.; Boulesteix, A.; Zeileis, A.; Hothorn, T. Bias in random forest variable importance measures: Illustrations, sources and a solution. BMC Bioinform. 2007, 8, 25. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Strobl, C.; Zeileis, A. Exploring the statistical properties of a test for random forest variable importance. In Compstat 2008—Proceedings in Computational Statistics, Porto, Portugal, 30 January 2008.
- Díaz-Uriarte, R.; De Andres, S.A. Gene selection and classification of microarray data using random forest. BMC Bioinform. 2006, 7, 3. [Google Scholar] [CrossRef] [PubMed]
- Kursa, M.B.; Rudnicki, W.R. Feature selection with the boruta package. J. Statis. Soft. 2010, 36, 1–13. [Google Scholar]
- Vincenzi, S.; Zucchetta, M.; Franzoi, P.; Pellizzato, M.; Pranovi, F.; De Leo, G.A.; Torricelli, P. Application of a random forest algorithm to predict spatial distribution of the potential yield of ruditapes philippinarum in the Venice lagoon, Italy. Ecol. Model. 2011, 222, 1471–1478. [Google Scholar] [CrossRef]
- Abdel-Rahman, E.M.; Ahmed, F.B.; Ismail, R. Random forest regression for sugarcane yield prediction in umfolozi, South Africa based on landsat TM and ETM+ derived spectral vegetation indices. In Sugarcane: Production, Cultivation and Uses; Goncalves, J.F., Correia, K.D., Eds.; NOVA Science Publishers, Inc: Hauppauge NY, USA, 2012; pp. 257–284. [Google Scholar]
- Abdel-Rahman, E.M.; Ahmed, F.B.; Ismail, R. Random forest regression and spectral band selection for estimating sugarcane leaf nitrogen concentration using EO-1 hyperion hyperspectral data. Int. J. Remote Sens. 2013, 34, 712–728. [Google Scholar] [CrossRef]
- Adjorlolo, C.; Mutanga, O.; Cho, M.A.; Ismail, R. Spectral resampling based on user-defined inter-band correlation filter: C3 and C4 grass species classification. Int. J. Appl. Earth Obs. Geoinf. 2013, 21, 535–544. [Google Scholar] [CrossRef]
- Mather, P.; Tso, B. Classification Methods for Remotely senSed Data; CRC press/Taylor and Francis Group: London, UK, 2003. [Google Scholar]
- Asner, G.P.; Jones, M.O.; Martin, R.E.; Knapp, D.E.; Hughes, R.F. Remote sensing of native and invasive species in Hawaiian forests. Remote Sens. Environ. 2008, 112, 1912–1926. [Google Scholar] [CrossRef]
- Shouse, M.; Liang, L.; Fei, S. Identification of understory invasive exotic plants with remote sensing in urban forests. Int. J. Appl. Earth Obs. Geoinf. 2013, 21, 525–534. [Google Scholar] [CrossRef]
- Amaral, C.H.; Roberts, D.A.; Almeida, T.I.; Souza Filho, C.R. Mapping invasive species and spectral mixture relationships with neotropical woody formations in southeastern Brazil. ISPRS J. Photogramm. Remote Sens. 2015, 108, 80–93. [Google Scholar] [CrossRef]
- Wise, R.M.; Wilgen, B.W.; Le Maitre, D.C. Costs, benefits and management options for an invasive alien tree species: The case of mesquite in the northern cape, South Africa. J. Arid Environ. 2012, 84, 80–90. [Google Scholar] [CrossRef]
- Shiferaw, H.; Teketay, D.; Nemomissa, S.; Assefa, F. Some biological characteristics that foster the invasion of prosopis juliflora (SW.) dc. At middle awash rift valley area, north-eastern Ethiopia. J. Arid Environ. 2004, 58, 135–154. [Google Scholar] [CrossRef]
- Kumar, L.; Dury, S.J.; Schmidt, K.; Skidmore, A. Imaging spectrometry and vegetation science. In Image Spectrometry; Meer, F.D.d., Jong, S.M.D., Eds.; Kluwer Academic Publishers: London, UK, 2003; Volume 3, pp. 111–156. [Google Scholar]
- Adam, E.; Mutanga, O. Spectral discrimination of papyrus vegetation (cyperus papyrus l.) in swamp wetlands using field spectrometry. ISPRS J. Photogramm. Remote Sens. 2009, 64, 612–620. [Google Scholar] [CrossRef]
- Chan, J.C.; Paelinckx, D. Evaluation of random forest and adabboost tree based ensembles classification and spectral band selection for ecotope mapping airborne hyperspectral imagery. Remote Sens. Environ. 2008. [Google Scholar] [CrossRef]
- Zhang, G.; Li, H.; Fang, B. Discriminating acidic and alkaline enzymes using a random forest model with secondary structure amino acid composition. Proc. Biochem. 2009, 44, 654–660. [Google Scholar] [CrossRef]
- Adam, E.; Mutanga, O.; Ismail, R. Determining the susceptibility of eucalyptus nitens forests to coryphodema tristis (cossid moth) occurrence in mpumalanga, South Africa. Int. J. Geogr. Inf. Sci. 2013, 27, 1–15. [Google Scholar] [CrossRef]
- Mansour, K.; Mutanga, O.; Everson, T.; Adam, E. Discriminating indicator grass species for rangeland degradation assessment using hyperspectral data resampled to Aisa eagle resolution. ISPRS J. Photogramm. Remote Sens. 2012, 70, 56–65. [Google Scholar] [CrossRef]
- Ismail, R.; Mutanga, O. Discriminating the early stages of sirex noctilio infestation using classification tree ensembles and shortwave infrared bands. Int. J. Remote Sens. 2011, 32, 4249–4266. [Google Scholar] [CrossRef]
- Adam, E.; Mutanga, O.; Abdel-Rahman, E.M.; Ismail, R. Estimating standing biomass in papyrus (cyperus papyrus l.) swamp: Exploratory of in situ hyperspectral indices and random forest regression. Int. J. Remote Sens. 2014, 35, 693–714. [Google Scholar] [CrossRef]
- Ceccato, P.; Flasse, S.; Tarantola, S.; Jacquemoud, S.; Grégoire, J. Detecting vegetation leaf water content using reflectance in the optical domain. Remote Sens. Environ. 2001, 77, 22–33. [Google Scholar] [CrossRef]
- Carter, G. Ratios of leaf reflectances in narrow wavebands as indicators of plant stress. Int. J. Remote Sens. 1994, 15, 697–703. [Google Scholar] [CrossRef]
- Ghulam, A.; Qin, Q.; Teyip, T.; Li, Z.-L. Modified perpendicular drought index (MPDI): A real-time drought monitoring method. ISPRS J. Photogramm. Remote Sens. 2007, 62, 150–164. [Google Scholar] [CrossRef]
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mureriwa, N.; Adam, E.; Sahu, A.; Tesfamichael, S. Examining the Spectral Separability of Prosopis glandulosa from Co-Existent Species Using Field Spectral Measurement and Guided Regularized Random Forest. Remote Sens. 2016, 8, 144. https://0-doi-org.brum.beds.ac.uk/10.3390/rs8020144
Mureriwa N, Adam E, Sahu A, Tesfamichael S. Examining the Spectral Separability of Prosopis glandulosa from Co-Existent Species Using Field Spectral Measurement and Guided Regularized Random Forest. Remote Sensing. 2016; 8(2):144. https://0-doi-org.brum.beds.ac.uk/10.3390/rs8020144
Chicago/Turabian StyleMureriwa, Nyasha, Elhadi Adam, Anshuman Sahu, and Solomon Tesfamichael. 2016. "Examining the Spectral Separability of Prosopis glandulosa from Co-Existent Species Using Field Spectral Measurement and Guided Regularized Random Forest" Remote Sensing 8, no. 2: 144. https://0-doi-org.brum.beds.ac.uk/10.3390/rs8020144