Moving Towards Dynamic Ocean Management: How Well Do Modeled Ocean Products Predict Species Distributions?
Abstract
:1. Introduction
2. Methods
2.1. Study Area and Field Methods

2.2. Analytical Methods
2.2.1. Habitat Variables
2.2.2. Modeling Framework
2.2.3. Density Predictions
2.2.4. Model Comparison
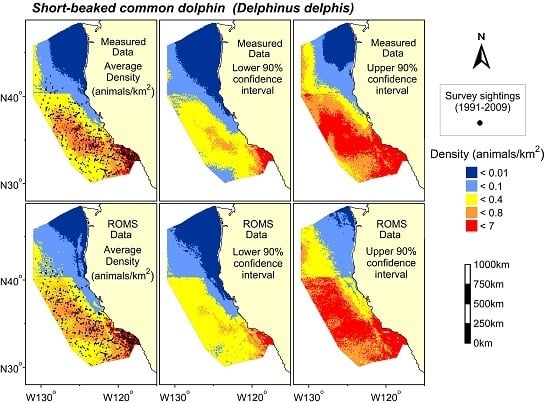
3. Results
| Species | Model type | Generalized Additive Model (GAM) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SST | MLD | lnCHL | SAL | sdSST | PEA | SSH | sdSSH | DEPTH | SLOPE | ASPECT | Bivariate | ||
| Striped dolphin | Measured | s(3.08)* | linear*** | ||||||||||
| ROMS | s(4.533)* | s(3.75)* | s(3.53)*** | ||||||||||
| Short-beaked common dolphin | Measured | s(4.64)*** | s(1.44)*** | s(5.69)*** | s(3.83)*** | s(5.47)*** | s(3.85)*** | ||||||
| ROMS | s(6.76)*** | s(5.80)*** | s(3.95)*** | s(6.72)*** | s(3.70)*** | ||||||||
| Long-beaked common dolphin | Measured | linear*** | s(2.92)** | linear** | te(LON,LAT, 5.65)*** | ||||||||
| ROMS | linear* | s(1.37)* | te(LON,LAT, 7.97)*** | ||||||||||
| Common bottlenose dolphin | Measured | s(3.91)** | s(2.85)* | te(LAT,d200, 12.46)*** | |||||||||
| ROMS | s(2.32)*** | s(3.41)** | linear* | linear** | te(LAT,d200, 12.22)*** | ||||||||
| Risso’s dolphin | Measured | s(4.46)* | te(LAT,d200, 17.49)*** | ||||||||||
| ROMS | s(6.70)** | s(1.59)* | te(LAT,d200, 17.63)*** | ||||||||||
| Pacific white-sided dolphin | Measured | s(4.89)** | s(3.44)*** | s(4.11)*** | |||||||||
| ROMS | linear** | s(3.63)*** | s(3.71)*** | ||||||||||
| Northern right whale dolphin | Measured | s(4.27)*** | linear*** | s(5.22)*** | |||||||||
| ROMS | s(3.64)*** | linear* | s(5.45)** | s(5.47)*** | |||||||||
| Dall’s porpoise | Measured | s(2.69)*** | s(5.54)*** | s(4.75)*** | s(5.43)*** | s(6.31)*** | linear** | ||||||
| ROMS | s(3.34)*** | s(5.32)*** | s(3.86)** | s(3.10)*** | linear* | s(6.56)*** | linear*** | ||||||
| Fin whale | Measured | s(4.68)*** | s(4.50)*** | s(6.96)*** | s(3.47)*** | s(2.65)*** | s(5.81)** | ||||||
| ROMS | s(4.24)*** | s(6.59)*** | s(4.75)** | s(3.92)*** | s(5.00)*** | te(LAT,MLD, 12.60)*** | |||||||
| Blue whale | Measured | s(5.85)*** | s(4.25)*** | s(7.02)*** | s(5.28)*** | s(4.96)** | |||||||
| ROMS | s(5.32)*** | s(3.56)*** | s(4.11)** | te(LAT,MLD, 13.73)*** | |||||||||
| Humpback whale | Measured | s(4.61)*** | s(3.90)*** | s(6.97)*** | s(1.64)*** | linear* | s(5.91)** | ||||||
| ROMS | s(6.41)*** | s(4.35)** | s(5.65)*** | s(4.97)*** | linear* | s(2.04)*** | linear* | s(6.76)*** | |||||
| Species | No. Sites | Data Type | Group Size Model | ||
|---|---|---|---|---|---|
| Measured | ROMS | ||||
| Striped dolphin | 103 | Expl.Dev. | 2.34 | 5.01 | 19.7 |
| RMSE | 0.086 | 0.086 | |||
| Ratio | 1.100 | 1.023 | |||
| Short-beaked common dolphin | 718 | Expl.Dev. | 5.50 | 2.79 | 8.4 |
| RMSE | 0.221 | 0.220 | |||
| Ratio | 1.004 | 1.049 | |||
| Long-beaked common dolphin | 128 | Expl.Dev. | 51.9 | 50.5 | 10.0 |
| RMSE | 0.089 | 0.088 | |||
| Ratio | 1.020 | 1.029 | |||
| Common bottlenose dolphin | 61 | Expl.Dev. | 31.9 | 34.5 | 15.3 |
| RMSE | 0.066 | 0.064 | |||
| Ratio | 0.992 | 1.000 | |||
| Risso’s dolphin | 172 | Expl.Dev. | 13.3 | 14.5 | 4.2 |
| RMSE | 0.110 | 0.109 | |||
| Ratio | 0.976 | 0.977 | |||
| Pacific white-sided dolphin | 137 | Expl.Dev. | 20.6 | 18.2 | 7.2 |
| RMSE | 0.097 | 0.099 | |||
| Ratio | 0.918 | 0.857 | |||
| Northern right whale dolphin | 108 | Expl.Dev. | 14.3 | 17.0 | 8.7 |
| RMSE | 0.085 | 0.084 | |||
| Ratio | 1.000 | 0.989 | |||
| Dall’s porpoise | 527 | Expl.Dev. | 37.0 | 36.0 | |
| RMSE | 1.041 | 1.043 | NA | ||
| Ratio | 0.939 | 0.944 | |||
| Fin whale | 362 | Expl.Dev. | 12.0 | 15.0 | |
| RMSE | 0.512 | 0.508 | NA | ||
| Ratio | 0.876 | 0.898 | |||
| Blue whale | 261 | Expl.Dev. | 16.3 | 17.3 | |
| RMSE | 0.258 | 0.257 | NA | ||
| Ratio | 0.953 | 0.953 | |||
| Humpback whale | 275 | Expl.Dev. | 48.3 | 47.0 | |
| RMSE | 0.322 | 0.337 | NA | ||
| Ratio | 0.98 | 0.974 | |||
| Model | Model | Model | Line-Transect | Line-Transect | |
|---|---|---|---|---|---|
| Species | Data Type | Grid Abundance | Grid Density | Segment Abundance | Segment Density |
| Striped dolphin | Measured | 18,310 | 0.016 | 19,887 | 0.017 |
| ROMS | 18,390 | 0.016 | |||
| Short-beaked common dolphin | Measured | 346,643 | 0.303 | 490,961 | 0.430 |
| ROMS | 351,523 | 0.308 | |||
| Long-beaked common dolphin | Measured | 60,476 | 0.053 | 161,589 | 0.142 |
| ROMS | 63,496 | 0.056 | |||
| Common bottlenose dolphin | Measured | 3,262 | 0.003 | 5,871 | 0.005 |
| ROMS | 12,455 | 0.011 | |||
| Risso’s dolphin | Measured | 13,840 | 0.012 | 14,935 | 0.013 |
| ROMS | 12,283 | 0.011 | |||
| Pacific white-sided dolphin | Measured | 36,774 | 0.032 | 33,446 | 0.029 |
| ROMS | 49,661 | 0.043 | |||
| Northern right whale dolphin | Measured | 24,671 | 0.022 | 20,302 | 0.018 |
| ROMS | 25,649 | 0.022 | |||
| Dall’s porpoise | Measured | 41,002 | 0.036 | 35,355 | 0.031 |
| ROMS | 42,827 | 0.037 | |||
| Fin whale | Measured | 4,337 | 0.004 | 4,322 | 0.004 |
| ROMS | 4,629 | 0.004 | |||
| Blue whale | Measured | 1,535 | 0.001 | 1,873 | 0.002 |
| ROMS | 1,805 | 0.002 | |||
| Humpback whale | Measured | 1,159 | 0.001 | 1,426 | 0.001 |
| ROMS | 1,319 | 0.001 |






4. Discussion
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Baumgartner, M.F.; Cole, T.V.N.; Clapham, P.J.; Mate, B.R. North Atlantic right whale habitat in the lower Bay of Fundy and on the SW Scotian Shelf during 1999–2001. Mar. Ecol. Prog. Ser. 2003, 264, 137–154. [Google Scholar] [CrossRef]
- Leathwick, J.R.; Elith, J.; Francis, M.P.; Hastie, T.; Taylor, P. Variation in demersal fish species richness in the oceans surrounding New Zealand: An analysis using boosted regression trees. Mar. Ecol. Prog. Ser. 2006, 321, 267–281. [Google Scholar] [CrossRef]
- Louzao, M.; Hyrenbach, K.D.; Arcos, J.; Abello, P.; Gil De Sola, L.; Oro, D. Oceanographic habitat of an endangered Mediterranean procellariiform: Implications for marine protected areas. Ecol. Appl. 2006, 16, 1683–1695. [Google Scholar] [CrossRef]
- Benson, S.R.; Eguchi, T.; Foley, D.G.; Forney, K.A.; Bailey, H.; Hitipeuw, C.; Samber, B.P.; Tapilatu, R.F.; Rei, V.; Ramohia, P.; et al. Large-scale movements and high-use areas of western Pacific leatherback turtles, Dermochelys coriacea. Ecosphere 2011. [Google Scholar] [CrossRef]
- Gilles, A.; Adler, S.; Kaschner, K.; Scheidat, M.; Siebert, U. Modelling harbor porpoise seasonal density as a function of the German Bight environment: Implications for management. Endanger. Species Res. 2011, 14, 157–169. [Google Scholar] [CrossRef]
- Goetz, K.T.; Montgomery, R.A.; Ver Hoef, J.M.; Hobbs, R.C.; Johnson, D.S. Identifying essential summer habitat of the endangered beluga whale Delphinapterus leucas in Cook Inlet, Alaska. Endanger. Species Res. 2012, 16, 135–147. [Google Scholar] [CrossRef]
- Keller, C.A.; Garrison, L.; Baumstark, R.; Ward-Geiger, L.I.; Hines, E. Application of a habitat model to define calving habitat of the North Atlantic right whale in the southeastern United States. Endanger. Species Res. 2012, 18, 73–87. [Google Scholar] [CrossRef]
- Oppel, S.; Meirinho, A.; Ramírez, I.; Gardner, B.; O’Connell, A.F.; Miller, P.I.; Louzao, M. Comparison of five modelling techniques to predict the spatial distribution and abundance of seabirds. Biol. Conserv. 2012, 156, 94–104. [Google Scholar] [CrossRef]
- Hammond, P.S.; Mcleod, K.; Berggren, P.; Borchers, D.L.; Burt, L.; Canadas, A.; Desportes, G.; Donovan, G.P.; Gilles, A.; Gillespie, R.G.; et al. Cetacean abundance and distribution in European Atlantic shelf waters to inform conservation and management. Biol. Conserv. 2013, 164, 107–122. [Google Scholar] [CrossRef] [Green Version]
- Redfern, J.V.; McKenna, M.F.; Moore, T.J.; Calambokidis, J.; DeAngelis, M.L.; Becker, E.A.; Barlow, J.; Forney, K.A.; Fiedler, P.C.; Chivers, S.J. Assessing the risk of ships striking large whales in marine spatial planning. Conserv. Biol. 2013, 27, 292–302. [Google Scholar] [CrossRef] [PubMed]
- Forney, K.A.; Becker, E.A.; Foley, D.G.; Barlow, J.; Oleson, E.M. Habitat-based models of cetacean density and distribution in the central North Pacific. Endanger. Species Res. 2015, 27, 1–20. [Google Scholar] [CrossRef]
- Torres, L.G.; Sutton, P.J.H.; Thompson, D.R.; Delord, K.; Weimerskirch, H.; Sagar, P.M.; Sommer, E.; Dilley, B.J.; Ryan, P.G.; Phillips, R.A. Poor transferability of species distribution models for a pelagic predator, the grey petrel, indicates contrasting habitat preferences across ocean basins. PLoS ONE 2015, 10, e0120014. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Becker, E.A.; Foley, D.G.; Forney, K.A.; Barlow, J.; Redfern, J.V.; Gentemann, C.L. Forecasting cetacean abundance patterns to enhance management decisions. Endanger. Species Res. 2012, 16, 97–112. [Google Scholar] [CrossRef]
- Shchepetkin, A.F.; McWilliams, J.C. The regional oceanic modeling system (ROMS): A split-explicit, free-surface, topography-following-coordinate oceanic model. Ocean Model. 2005, 9, 347–404. [Google Scholar] [CrossRef]
- Oke, P.R.; Brassington, G.B.; Griffin, D.A.; Schiller, A. The BLUElink ocean data assimilation system (BODAS). Ocean Model. 2008, 21, 46–70. [Google Scholar] [CrossRef]
- Chassignet, E.P.; Hurlburt, H.E.; Smedstad, O.M.; Halliwell, G.R.; Hogan, P.J.; Wallcraft, A.J.; Baraille, R.; Bleck, R. The HYCOM (Hybrid Coordinate Ocean Model) data assimilative system. J. Mar. Syst. 2007, 65, 60–83. [Google Scholar] [CrossRef]
- Barron, C.N.; Kara, A.B.; Martin, P.J.; Rhodes, R.C.; Smedstad, L.F. Formulation, implementation and examination of vertical coordinate choices in the Global Navy Coastal Ocean Model (NCOM). Ocean Model. 2006, 11, 347–375. [Google Scholar] [CrossRef]
- Carton, J.A.; Giese, B.S. A Reanalysis of ocean climate using Simple Ocean Data Assimilation (SODA). Mon. Weather Rev. 2008, 136, 2999–3017. [Google Scholar] [CrossRef]
- Moore, A.M.; Arango, H.G.; Broquet, G.; Edwards, C.A.; Veneziani, M.; Powell, B.S.; Foley, D.; Doyle, J.D.; Costa, D.; Robinson, P. The Regional Ocean Modeling System (ROMS) 4-dimensional variational data assimilation systems. II: Performance and application to the California current system. Prog. Oceanogr. 2011, 91, 50–73. [Google Scholar] [CrossRef]
- Edwards, C.A.; Moore, A.M.; Hoteit, I.; Cornuelle, B.D. Regional ocean data assimilation. Annu. Rev. Mar. Sci. 2015, 7, 21–42. [Google Scholar] [CrossRef] [PubMed]
- Fiedler, P.C.; Reilly, S.B. Interannual variability of dolphin habitats in the eastern tropical Pacific. I: Research vessel surveys, 1986–1990. Fish. Bull. 1994, 92, 434–450. [Google Scholar]
- Ballance, L.T.; Pitman, R.L.; Fiedler, P.C. Oceanographic influences on seabirds and cetaceans of the eastern tropical Pacific: A review. Prog. Oceanogr. 2006, 69, 360–390. [Google Scholar] [CrossRef]
- Ferguson, M.C.; Barlow, J.; Fiedler, P.; Reilly, S.B.; Gerrodette, T. Spatial models of delphinid (family Delphinidae) encounter rate and group size in the eastern tropical Pacific Ocean. Ecol. Model. 2006, 193, 645–662. [Google Scholar] [CrossRef]
- Forney, K.A.; Ferguson, M.C.; Becker, E.A.; Fiedler, P.C.; Redfern, J.V.; Barlow, J.; Vilchis, I.L.; Gerrodette, T.; Balance, L.T. Habitat-based spatial models of cetacean density in the eastern Pacific Ocean. Endanger. Species Res. 2012, 16, 113–133. [Google Scholar] [CrossRef]
- Dransfield, A.; Hines, E.; McGowan, J.; Holzman, B.; Nur, N.; Elliott, M.; Howar, J.; Jahncke, J. Where the whales are: Using habitat modeling to support changes in shipping regulations within National Marine Sanctuaries in Central California. Endanger. Species Res. 2014, 26, 39–57. [Google Scholar] [CrossRef]
- Hobday, A.J.; Hartog, J.R. Dynamic Ocean Features for use in Ocean Management. Oceanography 2014, 27, 134–145. [Google Scholar] [CrossRef]
- Lewison, R.L.; Hobday, A.J.; Maxwell, S.M.; Hazen, E.L.; Hartog, J.R.; Dunn, D.C.; Briscoe, D.K.; Fossette, S.; O’Keefe, C.E.; Barnes, M.; et al. Dynamic Ocean Management: Identifying the Critical Ingredients of Dynamic Approaches to Ocean Resource Management. Bioscience 2015. [Google Scholar] [CrossRef]
- Maxwell, S.M.; Hazen, E.L.; Lewison, R.L.; Dunn, D.C.; Bailey, H.; Bograd, S.J.; Briscoe, D.K.; Fossette, S.; Hobday, A.J.; Bennett, M.; et al. Dynamic ocean management: Defining and conceptualizing real-time management of the ocean. Mar. Policy 2015, 58, 42–50. [Google Scholar] [CrossRef]
- Hobday, A.J.; Hartmann, K. Near real-time spatial management based on habitat predictions for a longline bycatch species. Fish. Manag. Ecol. 2006, 13, 365–380. [Google Scholar] [CrossRef]
- Howell, E.A.; Kobayashi, D.R.; Parker, D.M.; Balazs, G.H.; Polovina, J.J. TurtleWatch: A tool to aid in the bycatch reduction of loggerhead turtles Caretta caretta in the Hawaii-based pelagic longline fishery. Endanger. Species Res. 2008, 5, 267–278. [Google Scholar] [CrossRef]
- Hobday, A.J.; Hartog, J.R.; Timmis, T.; Fielding, J. Dynamic spatial zoning to manage southern bluefin tuna capture in a multi-species longline fishery. Fish. Oceanogr. 2010, 19, 243–253. [Google Scholar] [CrossRef]
- Hartog, J.; Hobday, A.J.; Matear, R.; Feng, M. Habitat overlap of southern bluefin tuna and yellowfin tuna in the east coast longline fishery-implications for present and future spatial management. Deep Sea Res. Part II 2011, 58, 746–752. [Google Scholar] [CrossRef]
- Howell, E.A.; Hoover, A.A.; Benson, S.R.; Bailey, H.; Polovina, J.J.; Seminoff, J.A.; Dutton, P.H. Enhancing the TurtleWatch product for leatherback sea turtles, a dynamic habitat model for ecosystem-based management. Fish. Oceanogr. 2015, 24, 57–68. [Google Scholar] [CrossRef]
- Redfern, J.V.; Barlow, J.; Balance, L.T.; Gerrodette, T.; Becker, E.A. Absence of scale dependence in cetacean-habitat models for the eastern tropical Pacific Ocean. Mar. Ecol. Prog. Ser. 2008, 363, 1–14. [Google Scholar] [CrossRef]
- Barlow, J.; Ferguson, M.C.; Becker, E.A.; Redfern, J.V.; Forney, K.A.; Vilchis, I.L.; Fiedler, P.C.; Gerrodette, T.; Balance, L.T. Predictive Modeling of Cetacean Densities in the Eastern Pacific Ocean; NOAA Technical Memorandum NMFS-SWFSC-444; NOAA: La Jolla, CA, USA, 2009.
- Mannocci, L.; Catalogna, M.; Dorémus, G.; Laran, S.; Lehodey, P.; Massart, W.; Monestiez, P.; Van Canneyt, O.; Watremez, P.; Ridoux, V. Predicting cetacean and seabird habitats across a productivity gradient in the South Pacific gyre. Prog. Oceanogr. 2014, 120, 383–398. [Google Scholar] [CrossRef]
- Hazen, E.L.; Jorgensen, S.J.; Rykaczewski, R.; Bograd, S.J.; Foley, D.G.; Jonsen, I.D.; Shaffer, S.A.; Dunne, J.; Costa, D.P.; Crowder, L.B.; et al. Predicted Habitat Shifts of Pacific Top Predators in a Changing Climate. Nature Climate Change 2013. [Google Scholar] [CrossRef]
- Torres, L.G.; Smith, T.D.; Sutton, P.; Macdiarmid, A.; Bannister, J.; Miyashita, T. From exploitation to conservation: Habitat models using whaling data predict distribution patterns and threat exposure of an endangered whale. Divers. Distrib. 2013, 19, 1138–1152. [Google Scholar] [CrossRef]
- Becker, E.A.; Forney, K.A.; Foley, D.G.; Smith, R.C.; Moore, T.J.; Barlow, J. Predicting seasonal density patterns of California cetaceans based on habitat models. Endanger. Species Res. 2014, 23, 1–22. [Google Scholar] [CrossRef]
- Dell, J.T.; Wilcox, C.; Matear, R.J.; Chamberlain, M.A.; Hobday, A.J. Potential impacts of climate change on the distribution of longline catches of yellowfin tuna (Thunnus albacares) in the Tasman Sea. Deep Sea Res. Part II 2015, 113, 235–245. [Google Scholar] [CrossRef]
- Buckland, S.T.; Anderson, D.R.; Burnham, K.P.; Laake, J.L.; Borchers, D.L.; Thomas, L. Introduction to Distance Sampling: Estimating Abundance of Biological Populations; Oxford University Press: Oxford, UK, 2001. [Google Scholar]
- Barlow, J.; Forney, K.A. Abundance and density of cetaceans in the California Current ecosystem. Fish Bull. 2007, 105, 509–526. [Google Scholar]
- Carretta, J.V.; Chivers, S.J.; Perryman, W.L. Abundance of the long-beaked common dolphin (Delphinus capensis) in California and western Baja California waters estimated from a 2009 ship-based line-transect survey. Bull. South. Calif. Acad. Sci. 2011, 110, 152–164. [Google Scholar] [CrossRef]
- Kinzey, D.; Olson, P.; Gerrodette, T. Marine Mammal Data Collection Procedures on Research Ship Line-Transect Surveys by the Southwest Fisheries Science Center; Report No. LJ-00-08; Southwest Fisheries Science Center: La Jolla, CA, USA, 2000.
- Monterey, G.; Levitus, S. Seasonal Variability of Mixed Layer Depth for the World Ocean; NOAA Atlas NESDIS; NOAA: Silver Spring, MD, USA, 1997; Volume 14, p. 100.
- Holm-Hansen, O.; Lorenzen, C.J.; Holmes, R.W.; Strickland, J.D. Fluorometric determination of chlorophyll. J. Cons. Perm. Int. Explor. Mer. 1965, 30, 3–15. [Google Scholar] [CrossRef]
- Philbrick, V.A.; Fiedler, P.C.; Fluty, J.T.; Reilly, S.B. Report of Oceanographic Studies Conducted during the 2000 Eastern Tropical Pacific Ocean Survey on the Research Vessels David Starr Jordan and McArthur; NOAA Technical Memorandum NMFS-SWFSC-309; NOAA: La Jolla, CA, USA, 2001.
- Philbrick, V.A.; Fiedler, P.C.; Balance, L.T.; Demer, D.A. Report of Ecosystem Studies Conducted during the 2001 Oregon, California, and Washington (ORCAWALE) Marine Mammal Survey on the Research Vessels David Starr Jordan and McArthur; NOAA Technical Memorandum NMFS-SWFSC-349; NOAA: La Jolla, CA, USA, 2003.
- Reynolds, R.W.; Smith, T.M. Improved global sea surface temperature analyses using optimum interpolation. J. Clim. 1994, 7, 929–948. [Google Scholar] [CrossRef]
- Simons, R.A. ERDDAP. Available online: http://coastwatch.pfeg.noaa.gov/erddap (accessed on 26 February 2015).
- Becker, E.A.; Forney, K.A.; Ferguson, M.C.; Foley, D.G.; Smith, R.C.; Barlow, J.; Redfern, J.V. Comparing California Current cetacean-habitat models developed using in situ and remotely sensed sea surface temperature data. Mar. Ecol. Prog. Ser. 2010, 413, 163–183. [Google Scholar] [CrossRef]
- Crawford, W.; Moore, A.M.; Jacox, M.G.; Neveu, E.; Fiechter, J.; Edwards, C.A. An historical analysis of the California Current using ROMS 4D-Var. Part II: Climate variability. Ocean Model. in press.
- Neveu, E.; Moore, A.M.; Edwards, C.A.; Fiechter, J.; Drake, P.T.; Jacox, M.G.; Nuss, E. An historical analysis of the California Current using ROMS 4D-Var. Part I: System configuration and diagnostics. Ocean Model. in press. [CrossRef]
- Veneziani, M.; Edwards, C.A.; Doyle, J.D.; Foley, D.G. A central California coastal ocean modeling study: 1. Forward model and the influence of realistic versus climatological forcing. J. Geophys. Res. 2009, 114, C04015. [Google Scholar] [CrossRef]
- Broquet, G.; Edwards, C.A.; Moore, A.M.; Powell, B.S.; Veneziani, M.; Doyle, J.D. Application of 4D-Variational data assimilation to the California Current System. Dyn. Atmos. Oceans 2009. [Google Scholar] [CrossRef]
- Schroeder, I.D.; Santora, J.A.; Moore, A.M.; Edwards, C.A.; Fiechter, J.; Hazen, E.; Bograd, S.J.; Field, J.C.; Wells, B.K. Application of a data-assimilative regional ocean modeling system for assessing California Current System ocean conditions, krill, and juvenile rockfish interannual variability. Geophys. Res. Lett. 2014. [Google Scholar] [CrossRef]
- Amante, C.; Eakins, B.W. ETOPO1 1 arc-Minute Global Relief Model: Procedures, Data Sources and Analysis; Technical Memorandum NESDIS NGDC-24; National Geophysical Data Center: Boulder, CO, USA, 2009.
- Barlow, J.; Balance, L.T.; Forney, K.A. Effective Strip Widths for Ship-Based Line-Transect Surveys of Cetaceans; NOAA Technical Memorandum NMFS-SWFSC-484; NOAA: La Jolla, CA, USA, 2011.
- Hastie, T.J.; Tibshirani, R.J. Generalized Additive Models; Chapman & Hall/CRC: Boca Raton, FL, USA, 1990; Volume 43. [Google Scholar]
- Wood, S.N. Fast stable restricted maximum likelihood and marginal likelihood estimation of semiparametric Generalized Linear Models. J. R. Stat. Soc. Ser. B 2011, 73, 3–36. [Google Scholar] [CrossRef] [Green Version]
- Wood, S. Fast stable direct fitting and smoothness selection for generalized additive models. J. R. Statis. Soc. B 2008, 70, 495–518. [Google Scholar] [CrossRef] [Green Version]
- Marra, G.; Wood, S. Practical variable selection for generalized additive models. Comput. Stat. Data Anal. 2011, 55, 2372–2387. [Google Scholar] [CrossRef]
- Wood, S.N. Generalised Additive Models–An Introduction with R; Chapman and Hall/CRS: Boca Raton, FL, USA, 2006. [Google Scholar]
- Miller, D.L.; Burt, M.L.; Rexstad, E.A.; Thomas, L.; Gimenez, O. Spatial models for distance sampling data: Recent developments and future directions. Methods Ecol. Evol. 2013, 4, 1001–1010. [Google Scholar] [CrossRef]
- Cañadas, A.; Hammond, P.S. Abundance and habitat preferences of the short-beaked common dolphin Delphinus delphis in the southwestern Mediterranean: Implications for conservation. Endanger. Species Res. 2008, 4, 309–331. [Google Scholar] [CrossRef]
- Barlow, J. Inferring trackline detection probabilities, g(0), for cetaceans from apparent densities in different survey conditions. Mar. Mamm. Sci. 2015. [Google Scholar] [CrossRef]
- Wood, S.N. Thin plate regression splines. J. R. Stat. Soc. Ser. B 2003, 65, 95–114. [Google Scholar] [CrossRef]
- Reid, J.L., Jr.; Roden, G.I.; Wyllie, J.G. Studies of the California Current System. Calif. Ocean. Fish. Invest. Rep. 1958, 6, 28–56. [Google Scholar]
- McClatchie, S. Regional Fisheries Oceanography of the California Current System: The CalCOFI Program; Springer: New York, NY, USA, 2014. [Google Scholar]
- Forney, K.A. Environmental models of cetacean abundance: Reducing uncertainty in population trends. Conserv. Biol. 2000, 14, 1271–1286. [Google Scholar] [CrossRef]
- Moore, J.E.; Barlow, J. Bayesian state-space model of fin whale abundance trends from a 1991–2008 time series of line-transect surveys in the California Current. J. Appl. Ecol. 2011, 48, 1195–1205. [Google Scholar] [CrossRef]
- Barlow, J.; Calambokidis, J.; Falcone, E.A.; Baker, C.S.; Burdin, A.M.; Clapham, P.J.; Ford, J.K.B.; Gabriele, C.M.; LeDuc, R.; Mattila, D.K.; et al. Humpback whale abundance in the North Pacific estimated by photographic capture-recapture with bias correction from simulation studies. Mar. Mamm. Sci. 2011, 27, 793–818. [Google Scholar] [CrossRef]
- Becker, E.A.; Forney, K.A.; Foley, D.G.; Barlow, J. Density and Spatial Distribution Patterns of Cetaceans in the Central North Pacific Based on Habitat Models; NOAA Technical Memorandum NMFS-SWFSC-490; NOAA: La Jolla, CA, USA, 2012.
- Becker, E.A.; Forney, K.A.; Ferguson, M.C.; Barlow, J.; Redfern, J.V. Predictive Modeling of Cetacean Densities in the California Current Ecosystem Based on Summer/Fall Ship Surveys in 1991–2008; NOAA Technical Memorandum NMFS-SWFSC-499; NOAA: La Jolla, CA, USA, 2012.
- Calambokidis, J.; Steiger, G.H.; Curtice, C.; Harrison, J.; Ferguson, M.C.; Becker, E.A.; DeAngelis, M.; Van Parijs, S.M. Biologically important areas for selected cetaceans within U.S. waters-West Coast region. Aquat. Mamm. 2015, 41, 39–53. [Google Scholar] [CrossRef]
- Fiedler, P.C.; Reilly, S.B.; Hewitt, R.P.; Demer, D.; Philbrick, V.A.; Smith, S.; Armstrong, S.W.; Croll, D.A.; Tershy, B.R.; Mate, B.R. Blue whale habitat and prey in the California Channel Islands. Deep Sea Res. Part II Top. Stud. Oceanogr. 1998, 45, 1781–1801. [Google Scholar] [CrossRef]
- Heyning, J.E.; Perrin, W.F. Evidence for two species of common dolphin (genus Delphinus) from the eastern North Pacific. Contrib. Sci. Mus. Nat. Hist. Los Angel. 1994, 442, 1–35. [Google Scholar]
- Gerrodette, T.; Eguchi, T. Precautionary design of a marine protected area based on a habitat model. Endanger. Species Res. 2011, 15, 159–166. [Google Scholar] [CrossRef]
- Hobday, A.J.; Hartog, J.R.; Spillman, C.M.; Alves, O. Seasonal forecasting of tuna habitat for dynamic spatial management. Can. J. Fish. Aquat. Sci. 2011, 68, 898–911. [Google Scholar] [CrossRef]
- Hooker, S.K.; Whitehead, H.; Gowans, S. Marine protected area design and the spatial and temporal distribution of cetaceans in a submarine canyon. Conserv. Biol. 1999, 13, 592–602. [Google Scholar] [CrossRef]
- Cañadas, A.; Sagarminaga, R.; Urquiola, E.; Hammond, P.S. Habitat preference modelling as a conservation tool: Proposals for marine protected areas for cetaceans in southern Spanish waters. Aquat. Conserv. 2005, 15, 495–521. [Google Scholar] [CrossRef]
- Carretta, J.V.; Forney, K.A.; Laake, J.L. The abundance of southern California coastal bottlenose dolphins estimated from tandem aerial surveys. Mar. Mamm. Sci. 1998, 14, 655–675. [Google Scholar] [CrossRef]
- Defran, R.H.; Weller, D.W.; Kelly, D.L.; Espinosa, M.A. Range characteristics of Pacific coast bottlenose dolphins (Tursiops truncatus) in the Southern California Bight. Mar. Mamm. Sci. 1999, 15, 381–393. [Google Scholar] [CrossRef]
- Gregg, W.W. Assimilation of SeaWiFS ocean chlorophyll data into a three-dimensional global ocean model. J. Marine Syst. 2008, 69, 205–225. [Google Scholar] [CrossRef]
- Song, H.; Edwards, C.A.; Moore, A.M.; Fiechter, J. Data assimilation in a coupled physical-biogeochemical model of the California Current System using an incremental lognormal 4-dimensional variational approach: Part 1, Model formulation and biological data assimilation twin experiments. Ocean Model. submitted.
- Song, H.; Edwards, C.A.; Moore, A.M.; Fiechter, J. Data assimilation in a coupled physical-biogeochemical model of the California Current System using an incremental lognormal 4-dimensional variational approach: Part 2, Joint physical and biological data assimilation twin experiments. Ocean Model. in press.
- Song, H.; Edwards, C.A.; Moore, A.M.; Fiechter, J. Data assimilation in a coupled physical-biogeochemical model of the California Current System using an incremental lognormal 4-dimensional variational approach: Part 3, Assimilation in a realistic context using satellite and in situ observations. Ocean Model. in press.
- Croll, D.A.; Marinovic, B.B.; Benson, S.; Chavez, F.P.; Black, N.; Ternullo, R.; Tershy, B.R. From wind to whales: Trophic links in a coastal upwelling system. Mar. Ecol. Prog. Ser. 2005, 289, 117–130. [Google Scholar] [CrossRef]
- Barlow, J. Cetacean Abundance in the California Current Estimated from a 2008 Ship-Based Line-Transect Survey; NOAA Technical Memorandum NMFS-SWFSC-456; NOAA: La Jolla, CA, USA, 2010.
- Barlow, J. Cetacean Abundance in the California Current Estimated from Ship-Based Line-Transect Surveys in 1991–2014; NOAA Administrative Report; NMFS-SWFSC; NOAA: La Jolla, CA, USA, in press.
- Bond, N.A.; Cronin, M.F.; Freeland, H.; Mantua, N. Causes and impacts of the 2014 warm anomaly in the NE Pacific. Geophys. Res. Lett. 2015, 42, 3414–3420. [Google Scholar] [CrossRef]
- Hazen, E.L.; Nowacek, D.P.; St Laurent, L.; Halpin, P.N.; Moretti, D.J. The relationship among oceanography, prey fields, and beaked whale foraging habitat in the Tongue of the Ocean. PLoS ONE 2011, 6, e19269. [Google Scholar] [CrossRef] [PubMed]
- Lambert, C.; Mannocci, L.; Lehodey, P.; Ridoux, V. Predicting cetacean habitats from their energetic needs and the distribution of their prey in two contrasted tropical regions. PLoS ONE 2014, 9, e105958. [Google Scholar] [CrossRef] [PubMed]
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Becker, E.A.; Forney, K.A.; Fiedler, P.C.; Barlow, J.; Chivers, S.J.; Edwards, C.A.; Moore, A.M.; Redfern, J.V. Moving Towards Dynamic Ocean Management: How Well Do Modeled Ocean Products Predict Species Distributions? Remote Sens. 2016, 8, 149. https://0-doi-org.brum.beds.ac.uk/10.3390/rs8020149
Becker EA, Forney KA, Fiedler PC, Barlow J, Chivers SJ, Edwards CA, Moore AM, Redfern JV. Moving Towards Dynamic Ocean Management: How Well Do Modeled Ocean Products Predict Species Distributions? Remote Sensing. 2016; 8(2):149. https://0-doi-org.brum.beds.ac.uk/10.3390/rs8020149
Chicago/Turabian StyleBecker, Elizabeth A., Karin A. Forney, Paul C. Fiedler, Jay Barlow, Susan J. Chivers, Christopher A. Edwards, Andrew M. Moore, and Jessica V. Redfern. 2016. "Moving Towards Dynamic Ocean Management: How Well Do Modeled Ocean Products Predict Species Distributions?" Remote Sensing 8, no. 2: 149. https://0-doi-org.brum.beds.ac.uk/10.3390/rs8020149