Airborne Lidar Estimation of Aboveground Forest Biomass in the Absence of Field Inventory
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Site
2.2. Field Inventory
2.3. Lidar Inventory
2.3.1. Lidar Data Measurements
2.3.2. Forest Metrics Extraction
2.4. Aboveground Biomass Estimation Using Field Measurements
2.5. Aboveground Biomass Estimation Using Lidar Measurements
2.6. Aboveground Biomass Estimation Using Field and Lidar Measurements
3. Results and Discussion
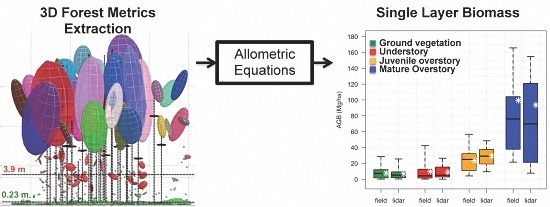
3.1. Aboveground Biomass at the Forest Layer Level
3.2. Aboveground Biomass at the Forest Plot Level
3.3. Aboveground Biomass at the Forest Plot Level Using a Regression Model Approach
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| UN-REDD | United Nations collaborative initiative on Reducing Emissions from Deforestation and forest Degradation |
| AGB | Aboveground biomass |
| AMS3D | 3D adaptive mean shift |
| bd | Bulk density |
| cbh | Crown base height |
| cc | Crown cover |
| CD | Correctly-detected trees |
| CDM | Canopy density models |
| dbh | Diameter at breast height |
| dh | Dominant height |
| ID | Incorrectly-detected trees |
| IQR | Inter-quartile range |
| GPS | Global positioning system |
| KDE | Kernel density estimators |
| MRV | Measuring, reporting and verification |
| th | Tree height |
| UD | Undetected trees |
| UNFCCC | United Nations Framework Convention on Climate Change |
| 3D | Three-dimensional |
References
- Zolkos, S.G.; Goetz, S.J.; Dubayah, R. A meta-analysis of terrestrial aboveground biomass estimation using lidar remote sensing. Remote Sens. Environ. 2013, 128, 289–298. [Google Scholar] [CrossRef]
- Houghton, R.; Greenglass, N.; Baccini, A.; Cattaneo, A.; Goetz, S.; Kellndorfer, J.; Laporte, N.; Walker, W. The role of science in Reducing Emissions from Deforestation and Forest Degradation (REDD). Carbon Manag. 2010, 1, 253–259. [Google Scholar] [CrossRef]
- Hall, F.G.; Bergen, K.; Blair, J.B.; Dubayah, R.; Houghton, R.; Hurtt, G.; Kellndorfer, J.; Lefsky, M.; Ranson, J.; Saatchi, S.; et al. Characterizing 3D vegetation structure from space: Mission requirements. Remote Sens. Environ. 2011, 115, 2753–2775. [Google Scholar] [CrossRef]
- Næsset, E. Predicting forest stand characteristics with airborne scanning laser using a practical two-stage procedure and field data. Remote Sens. Environ. 2002, 80, 88–99. [Google Scholar] [CrossRef]
- McRoberts, R.E.; Tomppo, E.O. Remote sensing support for national forest inventories. Remote Sens. Environ. 2007, 110, 412–419. [Google Scholar] [CrossRef]
- Meyer, V.; Saatchi, S.S.; Chave, J.; Dalling, J.W.; Bohlman, S.; Fricker, G.A.; Robinson, C.; Neumann, M.; Hubbell, S. Detecting tropical forest biomass dynamics from repeated airborne lidar measurements. Biogeosciences 2013, 10, 5421–5438. [Google Scholar] [CrossRef]
- Frazer, G.W.; Magnussen, S.; Wulder, M.A.; Niemann, K.O. Simulated impact of sample plot size and co-registration error on the accuracy and uncertainty of lidar-derived estimates of forest stand biomass. Remote Sens. Environ. 2011, 115, 636–649. [Google Scholar] [CrossRef]
- Mauya, E.; Hansen, E.; Gobakken, T.; Bollandsås, O.; Malimbwi, R.; Næsset, E. Effects of field plot size on prediction accuracy of aboveground biomass in airborne laser scanning-assisted inventories in tropical rain forests of Tanzania. Carbon Balance Manag. 2015, 10, 10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saatchi, S.; Mascaro, J.; Xu, L.; Keller, M.; Yang, Y.; Duffy, P.; Espírito-Santo, F.; Baccini, A.; Chambers, J.; Schimel, D. Seeing the forest beyond the trees. Glob. Ecol. Biogeogr. 2015, 24, 606–610. [Google Scholar] [CrossRef]
- Mitchard, E.T.A.; Saatchi, S.S.; Baccini, A.; Asner, G.P.; Goetz, S.J.; Harris, N.L.; Brown, S. Uncertainty in the spatial distribution of tropical forest biomass: A comparison of pan-tropical maps. Carbon Balance Manag. 2013, 8, 10. [Google Scholar] [CrossRef] [PubMed]
- Saatchi, S.S.; Harris, N.L.; Brown, S.; Lefsky, M.; Mitchard, E.T.A.; Salas, W.; Zutta, B.R.; Buermann, W.; Lewis, S.L.; Hagen, S.; et al. Benchmark map of forest carbon stocks in tropical regions across three continents. Proc. Natl. Acad. Sci. USA 2011, 108, 9899–9904. [Google Scholar] [CrossRef] [PubMed]
- Wulder, M.A.; White, J.C.; Nelson, R.F.; Næsset, E.; Ørka, H.O.; Coops, N.C.; Hilker, T.; Bater, C.W.; Gobakken, T. Lidar sampling for large-area forest characterization: A review. Remote Sens. Environ. 2012, 121, 196–209. [Google Scholar] [CrossRef]
- Ferraz, A.; Bretar, F.; Jacquemoud, S.; Gonçalves, G.; Pereira, L. 3D segmentation of forest structure using a mean-shift based algorithm. In Proceedings of the 17th IEEE International Conference on Image Processing (ICIP), Hong Kong, China, 26–29 September 2010; pp. 1413–1416.
- Ferraz, A.; Bretar, F.; Jacquemoud, S.; Gonçalves, G.; Pereira, L.; Tomé, M.; Soares, P. 3-D mapping of a multi-layered Mediterranean forest using ALS data. Remote Sens. Environ. 2012, 121, 210–223. [Google Scholar] [CrossRef]
- Ferraz, A.; Mallet, C.; Jacquemoud, S.; Gonçalves, G.R.; Tomé, M.; Soares, P.; Pereira, L.G.; Bretar, F. Canopy density model: A new ALS-derived product to generate multilayer crown cover maps. IEEE Trans. Geosci. Remote Sens. 2015, 53, 6776–6790. [Google Scholar] [CrossRef]
- Ferraz, A.; Saatchi, S.; Mallet, C.; Meyer, V. Lidar detection of individual tree size in tropical forests. Remote Sens. Environ. 2016, 183, 318–333. [Google Scholar] [CrossRef]
- Autoridade Florestal Nacional (AFN). In Instruçoes para o Trabalho de Campo do Inventario Florestal Nacional. Divisao para a Intervençao Florestal, Autoridade Florestal Nacional; Direcçao de Unidade de Gestao Florestal, Divisao para a Intervençao Florestal: Lisboa, Portugal, 2009.
- Stokes, J.; Ashmore, C.; Rawlins, L.; Sirois, L. Glossary of Terms Used in Timber Harvesting and Forest Engineering; General Technical Report SO-73; Forest Service, Southern Forest Experiment Station: New Orleans, LA, USA, 1989. [Google Scholar]
- Gonsamo, A.; D’odorico, P.; Pellikka, P. Measuring fractional forest canopy element cover and openness—Definitions and methodologies revisited. Oikos 2013, 122, 1283–1291. [Google Scholar] [CrossRef]
- Gonçalves, G.; Pereira, L. A thorough accuracy estimation of DTM produced from airborne full-waveform laser scanning data of unmanaged Eucalyptus plantations. IEEE Trans. Geosci. Remote Sens. 2012, 50, 3256–3266. [Google Scholar] [CrossRef]
- Khachiyan, L. Rounding of polytopes in the real number model of computation. Math. Oper. Res. 1996, 21, 307–320. [Google Scholar] [CrossRef]
- Riaño, D.; Chuvieco, E.; Ustin, S.L.; Salas, J.; Rodríguez-Pérez, J.R.; Ribeiro, L.M.; Viegas, D.X.; Moreno, J.M.; Fernández, H. Estimation of shrub height for fuel-type mapping combining airborne lidar and simultaneous color infrared ortho imaging. Int. J. Wildland Fire 2007, 16, 341–348. [Google Scholar] [CrossRef]
- António, N.; Tomé, M.; Tomé, J.; Soares, P.; Fontes, L. Effect of tree, stand, and site variables on the allometry of Eucalyptus globulus tree biomass. Can. J. For. Res. 2007, 37, 895–906. [Google Scholar] [CrossRef]
- Simões, S. Expansão ao Alentejo e Algarve de uma Curva de Acumulação Pós-Fogo Para a Biomassa Arbustiva. Master’s Thesis, Universidade Técnica de Lisboa, Instituto Superior de Agronomia, Lisboa, Portugal, 2006. [Google Scholar]
- Soares, P.; Tomé, M. Airborne laser scanning technologies—Need to estimate tree variables normally obtained in traditional forest inventory. In Proceedings of IUFRO Conference on Mixed and Pure Forest in a Changing World, Vila Real, Portugal, 8–10 October 2010.
- Popescu, S. Estimating biomass of individual pine trees using airborne lidar. Biomass Bioenergy 2007, 31, 646–655. [Google Scholar] [CrossRef]
- Silva, C.; Klauberg, C.; Carvalho, S.; Hudak, A.; Rodriguez, L. Mapping abouveground carbon stocks using lidar data in Eucalyptus spp in the state of São Paulo, Brazil. Sci. For. 2014, 42, 591–604. [Google Scholar]
- R Development Core Team. R: A Language Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2015. [Google Scholar]
- Dalponte, M.; Bruzzone, L.; Gianelle, D. Tree species classification in the Southern Alps based on the fusion of very high geometrical resolution multispectral/hyperspectral images and lidar data. Remote Sens. Environ. 2012, 123, 258–270. [Google Scholar] [CrossRef]
- Gachet, S.; Vela, E.; Tatoni, T. BASECO: A floristic and ecological database of Mediterranean French flora. Biodivers. Conserv. 2005, 14, 1023–1034. [Google Scholar] [CrossRef]
- Hazen, H. Biodiversity mapping. In International Encyclopedia of Human Geography; Rob, K., Nigel, T., Eds.; Elsevier: Oxford, UK, 2009; pp. 314–319. [Google Scholar]
- García, M.; Riaño, D.; Chuvieco, E.; Danson, F. Estimating biomass carbon stocks for a Mediterranean forest in central Spain using lidar height and intensity data. Remote Sens. Environ. 2010, 114, 816–830. [Google Scholar] [CrossRef]
- Sandberg, D.V.; Ottmar, R.D.; Cushon, G.H. Characterizing fuels in the 21st Century. Int. J. Wildland Fire 2001, 10, 381–387. [Google Scholar] [CrossRef]
- Anderson, H. Aids to Determining Fuels Models for Estimating Fire Behavior; Department of Agriculture, Forest Service, Rocky Mountain Research Station: Ogden, UT, USA, 1982.
- Finney, M. FARSITE: Fire Area Simulator-Model Development and Evaluation; Department of Agriculture, Forest Service, Rocky Mountain Research Station: Ogden, UT, USA, 2004.
- Andrews, P.L. Current status and future needs of the BehavePlus Fire Modeling System. Int. J. Wildland Fire 2014, 23, 21–33. [Google Scholar] [CrossRef]
- Fernandes, P.; Luz, A.; Loureiro, C.; Ferreira-Godinho, P.; Botelho, H. Fuel modelling and fire hazard assessment based on data from the Portuguese National Forest Inventory. For. Ecol. Manag. 2006, 234, S229. [Google Scholar] [CrossRef]
- Houghton, R.; Hall, F.; Goetz, S. Importance of biomass in the global carbon cycle. J. Geophys. Res. 2009, 114, 2156–2202. [Google Scholar] [CrossRef]
- Duncanson, L.I.; Cook, B.D.; Hurtt, G.C.; Dubayah, R.O. An efficient, multi-layered crown delineation algorithm for mapping individual tree structure across multiple ecosystems. Remote Sens. Environ. 2014, 154, 378–386. [Google Scholar] [CrossRef]
- Paris, C.; Member, S.; Valduga, D.; Bruzzone, L. A hierarchical approach to three-dimensional segmentation of lidar data at single-tree level in a multilayered forest. IEEE Trans. Geosci. Remote Sens. 2016, 54, 1–14. [Google Scholar] [CrossRef]
- Jakubowski, M.; Li, W.; Guo, Q.; Kelly, M. Delineating individual trees from lidar data: A comparison of vector- and raster-based segmentation approaches. Remote Sens. 2013, 5, 4163–4186. [Google Scholar] [CrossRef]
- Chave, J.; Réjou-Méchain, M.; Búrquez, A.; Chidumayo, E.; Colgan, M.; Delitti, W.; Duque, A.; Eid, T.; Fearnside, P.; Goodman, R.; et al. Improved allometric models to estimate the aboveground biomass of tropical trees. Glob. Chang. Biol. 2014, 20, 3177–3190. [Google Scholar] [CrossRef] [PubMed]
- Dechesne, C.; Mallet, C.; Le Bris, A.; Hervieu, A.; Gouet-Brunet, V. Forest stand segmentation using airborne lidar data and very high resolution multispectral imagery. ISPRS Arch. Photogramm. Remote Sens. Spat. Inf. Sci. 2016, XLI-B3, 207–214. [Google Scholar] [CrossRef]





| AGB (kg) | |||||
| Individual trees | Stem | if | (1) | ||
| if | |||||
| Bark | if | (2) | |||
| if | |||||
| Leaves | if | (3) | |||
| if | |||||
| Branches | if | (4) | |||
| if | |||||
| Total | (5) | ||||
| Forest layers | (6) | ||||
| dbh (cm) | |||||
| Individual trees | (7) | ||||
| n | R2 | RMSE Mg·ha−1 | RMSE (%) | Bias Mg·ha−1 | Bias (%) | |
|---|---|---|---|---|---|---|
| Single layer level | ||||||
| Mature overstory | 30 | 0.99 | 18 | 18.1 | −5.8 | 5.9 |
| Juvenile overstory | 10 | 0.38 | 13.3 | 56.7 | +5.8 | 24 |
| Understory | 30 | 0.37 | 9.9 | 101.3 | −0.8 | 8.9 |
| Ground vegetation | 40 | 0.65 | 4.1 | 53.3 | −0.7 | 9.5 |
| Forest plot level | ||||||
| Forest plot | 40 | 0.99 | 16.3 | 17.1 | −4.4 | 4.6 |
| Forest plot level using a traditional regression model approach | ||||||
| Forest plot* | 40 | 0.55 | 103.2 | 107.6 | −9.4 | 9.9 |
| Forest plot** | 39 | 0.72 | 23.32 | 31.1 | 0.1 | 0.1 |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ferraz, A.; Saatchi, S.; Mallet, C.; Jacquemoud, S.; Gonçalves, G.; Silva, C.A.; Soares, P.; Tomé, M.; Pereira, L. Airborne Lidar Estimation of Aboveground Forest Biomass in the Absence of Field Inventory. Remote Sens. 2016, 8, 653. https://0-doi-org.brum.beds.ac.uk/10.3390/rs8080653
Ferraz A, Saatchi S, Mallet C, Jacquemoud S, Gonçalves G, Silva CA, Soares P, Tomé M, Pereira L. Airborne Lidar Estimation of Aboveground Forest Biomass in the Absence of Field Inventory. Remote Sensing. 2016; 8(8):653. https://0-doi-org.brum.beds.ac.uk/10.3390/rs8080653
Chicago/Turabian StyleFerraz, António, Sassan Saatchi, Clément Mallet, Stéphane Jacquemoud, Gil Gonçalves, Carlos Alberto Silva, Paula Soares, Margarida Tomé, and Luisa Pereira. 2016. "Airborne Lidar Estimation of Aboveground Forest Biomass in the Absence of Field Inventory" Remote Sensing 8, no. 8: 653. https://0-doi-org.brum.beds.ac.uk/10.3390/rs8080653