Biodiversity Monitoring in Changing Tropical Forests: A Review of Approaches and New Opportunities
Abstract
:1. Introduction
- Give an overview of the state of the art and synthesise previous research on biodiversity monitoring in the context of changing tropical forests;
- Assess the potential of using evolving technologies and tools to further increase the detail and accuracy of biodiversity monitoring;
- Identify remaining gaps and opportunities on biodiversity monitoring approaches through evaluating their contribution to addressing the primary biodiversity attributes according to Noss [39];
- Assess how evolving technologies can help operationalise relevant EBVs for tropical forest environments.
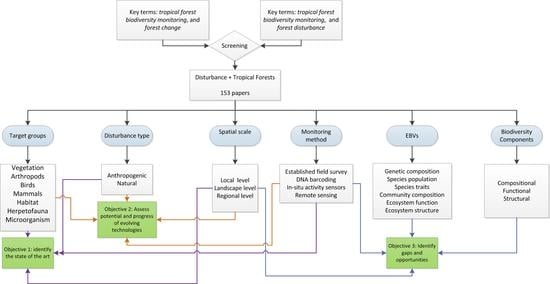
2. Analytical Framework and Data Analysis
3. Results
3.1. Spatial Scale
3.2. Disturbance Types
Disturbance Types per Country
3.3. Targeted Groups in Monitoring of Disturbed Tropical Forests
3.4. Monitoring Approaches over Time
Monitoring Approaches vs. Biodiversity Estimation Significance Values
3.5. Recent Technologies and New Opportunities for EBVs
4. Discussion
4.1. State of the Art
4.2. Potential and Progress of Evolving Technologies
4.3. Gaps and Opportunities
4.3.1. Monitoring of Primary Biodiversity Attributes
4.3.2. Operationalising EBVs with State-of-the-Art Technologies
5. Future Directions and Recommendations
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Barlow, J.; Lennox, G.D.; Ferreira, J.; Berenguer, E.; Lees, A.C.; Mac Nally, R.; Thomson, J.R.; de Barros Ferraz, S.F.; Louzada, J.; Oliveira, V.H.F. Anthropogenic disturbance in tropical forests can double biodiversity loss from deforestation. Nature 2016, 535, 144–147. [Google Scholar] [CrossRef] [PubMed]
- Turnhout, E.; Gupta, A.; Weatherley-Singh, J.; Vijge, M.J.; De Koning, J.; Visseren-Hamakers, I.J.; Herold, M.; Lederer, M. Envisioning redd+ in a post-paris era: Between evolving expectations and current practice. Wiley Interdiscip. Rev. Clim. Chang. 2016. [Google Scholar] [CrossRef]
- Thomas, S.C.; Baltzer, J.L. Tropical forests. In eLS; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2001. [Google Scholar]
- Bonan, G.B. Forests and climate change: Forcings, feedbacks, and the climate benefits of forests. Science 2008, 320, 1444–1449. [Google Scholar] [CrossRef] [PubMed]
- Malhi, Y.; Gardner, T.A.; Goldsmith, G.R.; Silman, M.R.; Zelazowski, P. Tropical forests in the anthropocene. In Annual Review of Environment and Resources; Gadgil, A., Liverman, D.M., Eds.; Annual Reviews: Palo Alto, CA, USA, 2014; Volume 39, pp. 125–159. [Google Scholar]
- Herold, M.; Román-Cuesta, R.; Heymell, V.; Hirata, Y.; Van Laake, P.; Asner, G.; Souza, C.; Avitabile, V.; MacDicken, K. A review of methods to measure and monitor historical carbon emissions from forest degradation. Unasylva 2011, 62, 238. [Google Scholar]
- Romijn, E.; Lantican, C.B.; Herold, M.; Lindquist, E.; Ochieng, R.; Wijaya, A.; Murdiyarso, D.; Verchot, L. Assessing change in national forest monitoring capacities of 99 tropical countries. For. Ecol. Manag. 2015, 352, 109–123. [Google Scholar] [CrossRef]
- Scholes, R.J.; Walters, M.; Turak, E.; Saarenmaa, H.; Heip, C.H.; Tuama, É.Ó.; Faith, D.P.; Mooney, H.A.; Ferrier, S.; Jongman, R.H. Building a global observing system for biodiversity. Curr. Opin. Environ. Sustain. 2012, 4, 139–146. [Google Scholar] [CrossRef]
- Vié, J.-C.; Hilton-Taylor, C.; Stuart, S.N. Wildlife in a Changing World: An Analysis of the 2008 IUCN Red List of Threatened Species; IUCN: Gland, Switerland, 2009. [Google Scholar]
- Convention on Biological Diversity (CBD). Decision X/2. In The Strategic Plan for Biodiversity 2011–2020 and the Aichi Biodiversity Targets 18 to 29 October 2010; Convention on Biological Diversity: Nagoya, Japan, 2010. [Google Scholar]
- Convention on Biological Diversity (CBD). Handbook of the Convention on Biological Diversity; Earthscan: London, UK, 2001; Volume 1. [Google Scholar]
- Transforming Our World: The 2030 Agenda for Sustainable. Available online: https://sustainabledevelopment.un.org/post2015/transformingourworld (accessed on 26 June 2017).
- United Nations (UN). Available online: http://www.un.org/sustainabledevelopment/development-agenda/ (accessed on 26 June 2017).
- Pereira, H.M.; Ferrier, S.; Walters, M.; Geller, G.; Jongman, R.; Scholes, R.; Bruford, M.; Brummitt, N.; Butchart, S.; Cardoso, A. Essential biodiversity variables. Science 2013, 339, 277–278. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kissling, W.D.; Ahumada, J.A.; Bowser, A.; Fernandez, M.; Fernández, N.; García, E.A.; Guralnick, R.P.; Isaac, N.J.; Kelling, S.; Los, W. Building essential biodiversity variables (ebvs) of species distribution and abundance at a global scale. Biol. Rev. Camb. Philos. Soc. 2017. [Google Scholar] [CrossRef] [PubMed]
- Newbold, T.; Hudson, L.N.; Phillips, H.R.; Hill, S.L.; Contu, S.; Lysenko, I.; Blandon, A.; Butchart, S.H.; Booth, H.L.; Day, J. A global model of the response of tropical and sub-tropical forest biodiversity to anthropogenic pressures. Proc. Biol. Sci. 2014, 281, 20141371. [Google Scholar] [CrossRef] [PubMed]
- Lee, P.S.; Sing, K.W.; Wilson, J.J. Reading mammal diversity from flies: The persistence period of amplifiable mammal mtdna in blowfly guts (chrysomya megacephala) and a new DNA mini-barcode target. PLoS ONE 2015, 10, e0123871. [Google Scholar] [CrossRef] [PubMed]
- Thomsen, P.F.; Willerslev, E. Environmental DNA—An emerging tool in conservation for monitoring past and present biodiversity. Biol. Conserv. 2015, 183, 4–18. [Google Scholar] [CrossRef]
- Bush, A.; Sollmann, R.; Wilting, A.; Bohmann, K.; Cole, B.; Balzter, H.; Martius, C.; Zlinszky, A.; Calvignac-Spencer, S.; Cobbold, C.A. Connecting earth observation to high-throughput biodiversity data. Nat. Ecol. Evol. 2017, 1, 176. [Google Scholar] [CrossRef] [PubMed]
- Marvin, D.C.; Koh, L.P.; Lynam, A.J.; Wich, S.; Davies, A.B.; Krishnamurthy, R.; Stokes, E.; Starkey, R.; Asner, G.P. Integrating technologies for scalable ecology and conservation. Glob. Ecol. Conserv. 2016, 7, 262–275. [Google Scholar] [CrossRef]
- Pimm, S.L.; Alibhai, S.; Bergl, R.; Dehgan, A.; Giri, C.; Jewell, Z.; Joppa, L.; Kays, R.; Loarie, S. Emerging technologies to conserve biodiversity. Trends Ecol. Evol. 2015, 30, 685–696. [Google Scholar] [CrossRef] [PubMed]
- Lausch, A.; Bannehr, L.; Beckmann, M.; Boehm, C.; Feilhauer, H.; Hacker, J.M.; Heurich, M.; Jung, A.; Klenke, R.; Neumann, C.; et al. Linking earth observation and taxonomic, structural and functional biodiversity: Local to ecosystem perspectives. Ecol. Indic. 2016, 70, 317–339. [Google Scholar] [CrossRef]
- Lindenmayer, D.B.; Gibbons, P.; Bourke, M.; Burgman, M.; Dickman, C.R.; Ferrier, S.; Fitzsimons, J.; Freudenberger, D.; Garnett, S.T.; Groves, C. Improving biodiversity monitoring. Austral Ecol. 2012, 37, 285–294. [Google Scholar] [CrossRef]
- Kuenzer, C.; Ottinger, M.; Wegmann, M.; Guo, H.; Wang, C.; Zhang, J.; Dech, S.; Wikelski, M. Earth observation satellite sensors for biodiversity monitoring: Potentials and bottlenecks. Int. J. Remote Sens. 2014, 35, 6599–6647. [Google Scholar] [CrossRef]
- Rose, R.A.; Byler, D.; Eastman, J.R.; Fleishman, E.; Geller, G.; Goetz, S.; Guild, L.; Hamilton, H.; Hansen, M.; Headley, R. Ten ways remote sensing can contribute to conservation. Conserv. Biol. 2015, 29, 350–359. [Google Scholar] [CrossRef] [PubMed]
- Mildrexler, D.J.; Zhao, M.; Heinsch, F.A.; Running, S.W. A new satellite-based methodology for continental-scale disturbance detection. Ecol. Appl. 2007, 17, 235–250. [Google Scholar] [CrossRef]
- Butchart, S.H.; Walpole, M.; Collen, B.; Van Strien, A.; Scharlemann, J.P.; Almond, R.E.; Baillie, J.E.; Bomhard, B.; Brown, C.; Bruno, J. Global biodiversity: Indicators of recent declines. Science 2010, 328, 1164–1168. [Google Scholar] [CrossRef] [PubMed]
- Hansen, M.C.; Potapov, P.V.; Moore, R.; Hancher, M.; Turubanova, S.A.; Tyukavina, A.; Thau, D.; Stehman, S.V.; Goetz, S.J.; Loveland, T.R.; et al. High-resolution global maps of 21st-century forest cover change. Science 2013, 342, 850–853. [Google Scholar] [CrossRef] [PubMed]
- Schäfer, E.; Heiskanen, J.; Heikinheimo, V.; Pellikka, P. Mapping tree species diversity of a tropical montane forest by unsupervised clustering of airborne imaging spectroscopy data. Ecol. Indic. 2016, 64, 49–58. [Google Scholar] [CrossRef]
- Koh, L.; Wich, S. Dawn of drone ecology: Low-cost autonomous aerial vehicles for conservation. Trop. Conserv. Sci. 2012, 5, 121–132. [Google Scholar] [CrossRef] [Green Version]
- Calders, K.; Newnham, G.; Burt, A.; Murphy, S.; Raumonen, P.; Herold, M.; Culvenor, D.; Avitabile, V.; Disney, M.; Armston, J. Nondestructive estimates of above-ground biomass using terrestrial laser scanning. Methods Ecol. Evol. 2015, 6, 198–208. [Google Scholar] [CrossRef]
- Pettorelli, N.; Wegmann, M.; Skidmore, A.; Mücher, S.; Dawson, T.P.; Fernandez, M.; Lucas, R.; Schaepman, M.E.; Wang, T.; O’Connor, B. Framing the concept of satellite remote sensing essential biodiversity variables: Challenges and future directions. Remote Sens. Ecol. Conserv. 2016. [Google Scholar] [CrossRef]
- Riitters, K.; Wickham, J.; O’Neill, R.; Jones, K.B.; Smith, E. Global-scale patterns of forest fragmentation. Conserv. Ecol. 2000, 4, 3. [Google Scholar] [CrossRef]
- Riitters, K.; Wickham, J.; Costanza, J.K.; Vogt, P. A global evaluation of forest interior area dynamics using tree cover data from 2000 to 2012. Landsc. Ecol. 2016, 31, 137–148. [Google Scholar] [CrossRef] [Green Version]
- Kays, R.; Crofoot, M.C.; Jetz, W.; Wikelski, M. Terrestrial animal tracking as an eye on life and planet. Science 2015, 348, aaa2478. [Google Scholar] [CrossRef] [PubMed]
- Ji, Y.; Ashton, L.; Pedley, S.M.; Edwards, D.P.; Tang, Y.; Nakamura, A.; Kitching, R.; Dolman, P.M.; Woodcock, P.; Edwards, F.A. Reliable, verifiable and efficient monitoring of biodiversity via metabarcoding. Ecol. Lett. 2013, 16, 1245–1257. [Google Scholar] [CrossRef] [PubMed]
- Taberlet, P.; Coissac, E.; Pompanon, F.; Brochmann, C.; Willerslev, E. Towards next-generation biodiversity assessment using DNA metabarcoding. Mol. Ecol. 2012, 21, 2045–2050. [Google Scholar] [CrossRef] [PubMed]
- Turner, W. Sensing biodiversity. Science 2014, 346, 301–302. [Google Scholar] [CrossRef] [PubMed]
- Noss, R.F. Indicators for monitoring biodiversity: A hierarchical approach. Conserv. Biol. 1990, 4, 355–364. [Google Scholar] [CrossRef]
- CBD. What Is Forest Biological Diversity? Available online: http://www.fao.org/3/a-ac547e/y3582e02.htm (accessed on 26 June 2017).
- Whittaker, R.J.; Willis, K.J.; Field, R. Scale and species richness: Towards a general, hierarchical theory of species diversity. J. Biogeogr. 2001, 28, 453–470. [Google Scholar] [CrossRef]
- Beaudrot, L.; Ahumada, J.A.; O’Brien, T.; Alvarez-Loayza, P.; Boekee, K.; Campos-Arceiz, A.; Eichberg, D.; Espinosa, S.; Fegraus, E.; Fletcher, C.; et al. Standardized Assessment of Biodiversity Trends in Tropical Forest Protected Areas: The End Is Not in Sight. PLoS Biol. 2016, 14, e1002357. [Google Scholar] [CrossRef] [PubMed]
- Ellis, P.; Griscom, B.; Walker, W.; Goncalves, F.; Cormier, T. Mapping selective logging impacts in borneo with gps and airborne LiDAR. For. Ecol. Manag. 2016, 365, 184–196. [Google Scholar] [CrossRef]
- Rovero, F.; Martin, E.; Rosa, M.; Ahumada, J.A.; Spitale, D. Estimating species richness and modelling habitat preferences of tropical forest mammals from camera trap data. PLoS ONE 2014, 9, e103300. [Google Scholar] [CrossRef] [PubMed]
- Henle, K.; Potts, S.; Kunin, W.; Matsinos, Y.; Simila, J.; Pantis, J.; Grobelnik, V.; Penev, L.; Settele, J. Scaling in ecology and biodiversity conservation. Adv. Books 2014, 1, e1169. [Google Scholar]
- Rocchini, D. Seeing the unseen by remote sensing: Satellite imagery applied to species distribution modelling. J. Veg. Sci. 2013, 24, 209–210. [Google Scholar] [CrossRef]
- Pettorelli, N.; Laurance, W.F.; O’Brien, T.G.; Wegmann, M.; Nagendra, H.; Turner, W. Satellite remote sensing for applied ecologists: Opportunities and challenges. J. Appl. Ecol. 2014, 51, 839–848. [Google Scholar] [CrossRef]
- Haddad, N.M.; Brudvig, L.A.; Clobert, J.; Davies, K.F.; Gonzalez, A.; Holt, R.D.; Lovejoy, T.E.; Sexton, J.O.; Austin, M.P.; Collins, C.D.; et al. Habitat fragmentation and its lasting impact on earth’s ecosystems. Sci. Adv. 2015, 1, e1500052. [Google Scholar] [CrossRef] [PubMed]
- Fretwell, P.T.; LaRue, M.A.; Morin, P.; Kooyman, G.L.; Wienecke, B.; Ratcliffe, N.; Fox, A.J.; Fleming, A.H.; Porter, C.; Trathan, P.N. An emperor penguin population estimate: The first global, synoptic survey of a species from space. PLoS ONE 2012, 7, e33751. [Google Scholar] [CrossRef]
- Anderson, K.; Gaston, K.J. Lightweight unmanned aerial vehicles will revolutionize spatial ecology. Front. Ecol. Environ. 2013, 11, 138–146. [Google Scholar] [CrossRef]
- Blumstein, D.T.; Mennill, D.J.; Clemins, P.; Girod, L.; Yao, K.; Patricelli, G.; Deppe, J.L.; Krakauer, A.H.; Clark, C.; Cortopassi, K.A. Acoustic monitoring in terrestrial environments using microphone arrays: Applications, technological considerations and prospectus. J. Appl. Ecol. 2011, 48, 758–767. [Google Scholar] [CrossRef]
- Yu, X.; Wang, J.; Kays, R.; Jansen, P.A.; Wang, T.; Huang, T. Automated identification of animal species in camera trap images. EURASIP J. Image Video Process. 2013, 2013, 1–10. [Google Scholar] [CrossRef]
- Yu, D.W.; Ji, Y.; Emerson, B.C.; Wang, X.; Ye, C.; Yang, C.; Ding, Z. Biodiversity soup: Metabarcoding of arthropods for rapid biodiversity assessment and biomonitoring. Methods Ecol. Evol. 2012, 3, 613–623. [Google Scholar] [CrossRef]
- Hänfling, B.; Lawson Handley, L.; Read, D.S.; Hahn, C.; Li, J.; Nichols, P.; Blackman, R.C.; Oliver, A.; Winfield, I.J. Environmental DNA metabarcoding of lake fish communities reflects long-term data from established survey methods. Mol. Ecol. 2016. [Google Scholar] [CrossRef] [PubMed]
- Willerslev, E.; Davison, J.; Moora, M.; Zobel, M.; Coissac, E.; Edwards, M.E.; Lorenzen, E.D.; Vestergard, M.; Gussarova, G.; Haile, J.; et al. Fifty thousand years of arctic vegetation and megafaunal diet. Nature 2014, 506, 47–51. [Google Scholar] [CrossRef] [PubMed]
- Collins, R.; Cruickshank, R. The seven deadly sins of DNA barcoding. Mol. Ecol. Resour. 2013, 13, 969–975. [Google Scholar] [CrossRef] [PubMed]
- Proença, V.; Martin, L.J.; Pereira, H.M.; Fernandez, M.; McRae, L.; Belnap, J.; Böhm, M.; Brummitt, N.; García-Moreno, J.; Gregory, R.D. Global biodiversity monitoring: From data sources to essential biodiversity variables. Biol. Conserv. 2016, 213, 256–263. [Google Scholar] [CrossRef]
- Sanderson, J. Tropical ecology, assessment and monitoring initiative. Camera phototrapping monitoring protocol. Database 2015, 2015, bav054. [Google Scholar]
- Kattge, J.; Diaz, S.; Lavorel, S.; Prentice, I.C.; Leadley, P.; Bönisch, G.; Garnier, E.; Westoby, M.; Reich, P.B.; Wright, I.J. Try—A global database of plant traits. Philos. Trans. R. Soc. B Biol. Sci. 2010, 365, 3709–3718. [Google Scholar] [CrossRef] [Green Version]
- Baker, E.; Price, B.W.; Rycroft, S.D.; Hill, J.; Smith, V.S. Bioacoustica: A free and open repository and analysis platform for bioacoustics. Database 2015, 2015, bav054. [Google Scholar] [CrossRef] [PubMed]
- Morris, R.J. Anthropogenic impacts on tropical forest biodiversity: A network structure and ecosystem functioning perspective. Glob. Chang. Biol. 2011, 17, 2905–2935. [Google Scholar] [CrossRef] [PubMed]
- De Sy, V.; Herold, M.; Achard, F.; Asner, G.P.; Held, A.; Kellndorfer, J.; Verbesselt, J. Synergies of multiple remote sensing data sources for redd+ monitoring. Curr. Opin. Environ. Sustain. 2012, 4, 696–706. [Google Scholar] [CrossRef]
- Sexton, J.O.; Song, X.-P.; Feng, M.; Noojipady, P.; Anand, A.; Huang, C.; Kim, D.-H.; Collins, K.M.; Channan, S.; DiMiceli, C. Global, 30-m resolution continuous fields of tree cover: Landsat-based rescaling of modis vegetation continuous fields with LiDAR-based estimates of error. Int. J. Digit. Earth 2013, 6, 427–448. [Google Scholar] [CrossRef]
- Peres, C.A.; Barlow, J.; Laurance, W.F. Detecting anthropogenic disturbance in tropical forests. Trends Ecol. Evol. 2006, 21, 227–229. [Google Scholar] [CrossRef] [PubMed]
- Souza, J.L.P.; Baccaro, F.B.; Landeiro, V.L.; Franklin, E.; Magnusson, W.E.; Pequeno, P.A.C.L.; Fernandes, I.O. Taxonomic sufficiency and indicator taxa reduce sampling costs and increase monitoring effectiveness for ants. Divers. Distrib. 2016, 22, 111–122. [Google Scholar] [CrossRef]
- Mant, R.; Perry, E.; Heath, M.; Munroe, R.; Väänänen, E.; Großheim, C.; Kümper-Schlake, L. Addressing Climate Change—Why Biodiversity Matters; UNEP-WCMC: Cambridge, UK, 2014. [Google Scholar]
- Talbot, J.D. Carbon and Biodiversity Relationships in Tropical Forests. Available online: http://www.biotrade.org/congress/BackgroundDocs2/EI/UNREDD%20docs/Carbon%20&%20biodiversity%20relationships%20in%20tropical%20forests.pdf (accessed on 26 June 2017).
- Poorter, L.; Sande, M.; Thompson, J.; Arets, E.; Alarcón, A.; Álvarez-Sánchez, J.; Ascarrunz, N.; Balvanera, P.; Barajas-Guzmán, G.; Boit, A. Diversity enhances carbon storage in tropical forests. Glob. Ecol. Biogeogr. 2015, 24, 1314–1328. [Google Scholar] [CrossRef]
- Bustamante, M.; Roitman, I.; Aide, T.M.; Alencar, A.; Anderson, L.O.; Aragão, L.; Asner, G.P.; Barlow, J.; Berenguer, E.; Chambers, J. Toward an integrated monitoring framework to assess the effects of tropical forest degradation and recovery on carbon stocks and biodiversity. Glob. Chang. Biol. 2016, 22, 92–109. [Google Scholar] [CrossRef] [PubMed]
- Leblanc, G.; Francis, C.M.; Soffer, R.; Kalacska, M.; de Gea, J. Spectral reflectance of polar bear and other large arctic mammal pelts; potential applications to remote sensing surveys. Remote Sens. 2016, 8, 273. [Google Scholar] [CrossRef]
- Yang, Z.; Wang, T.; Skidmore, A.K.; de Leeuw, J.; Said, M.Y.; Freer, J. Spotting east african mammals in open savannah from space. PLoS ONE 2015, 9, e115989. [Google Scholar] [CrossRef] [PubMed]
- Thenkabail, P.S.; Lyon, J.G.; Huete, A. Hyperspectral Remote Sensing of Vegetation; CRC Press: Abingdon, UK, 2016. [Google Scholar]
- Badreldin, N.; Sanchez-Azofeifa, A. Estimating forest biomass dynamics by integrating multi-temporal landsat satellite images with ground and airborne LiDAR data in the coal valley mine, Alberta, Canada. Remote Sens. 2015, 7, 2832–2849. [Google Scholar] [CrossRef]
- Ioki, K.; Tsuyuki, S.; Hirata, Y.; Phua, M.H.; Wong, W.V.C.; Ling, Z.Y.; Johari, S.A.; Korom, A.; James, D.; Saito, H.; et al. Evaluation of the similarity in tree community composition in a tropical rainforest using airborne LiDAR data. Remote Sens. Environ. 2016, 173, 304–313. [Google Scholar] [CrossRef]
- O’Brien, T.G.; Baillie, J.E.M.; Krueger, L.; Cuke, M. The wildlife picture index: Monitoring top trophic levels. Anim. Conserv. 2010, 13, 335–343. [Google Scholar] [CrossRef]
- Peplow, M. Social sciences suffer from severe publication bias. Nature 2014. [Google Scholar] [CrossRef]
- Lortie, C.; Aarssen, L.; Budden, A.; Koricheva, J.; Leimu, R.; Tregenza, T. Publication bias and merit in ecology. Oikos 2007, 116, 1247–1253. [Google Scholar] [CrossRef]
- Pardieck, K.L.; Ziolkowski, D.J., Jr.; Lutmerding, M.; Campbell, K.; Hudson, M.-A.R. North American Breeding Bird Survey Dataset 1966–2015. Available online: https://www.mbr-pwrc.usgs.gov/bbs/BBS_Results_and_Analysis_2015.html (accessed on 26 June 2017).
- Sullivan, B.L.; Wood, C.L.; Iliff, M.J.; Bonney, R.E.; Fink, D.; Kelling, S. Ebird: A citizen-based bird observation network in the biological sciences. Biol. Conserv. 2009, 142, 2282–2292. [Google Scholar] [CrossRef]
- Dutch Phenological Network. Available online: https://www.naturetoday.com/intl/en/observations/natuurkalender (accessed on 26 June 2017).
- Nature and Biodiversity Conservation Union. Available online: https://www.nabu.de/imperia/md/content/nabude/nabu/jobs/nabu_agrobiodiv_study_ethiopia.pdf (accessed on 26 June 2017).
- Honrado, J.P.; Pereira, H.M.; Guisan, A. Fostering integration between biodiversity monitoring and modelling. J. Appl. Ecol. 2016, 53, 1299–1304. [Google Scholar] [CrossRef]
- GEO-BON. GEO BON Strategy for Development of Essential Biodiversity Variables. Available online: http://geobon.org/Downloads/Other_documents/Essential_Biodiversity_Variable_Strategy_v2.pdf (accessed on 26 June 2017).
- Skidmore, A.K.; Pettorelli, N. Agree on biodiversity metrics to track from space: Ecologists and space agencies must forge a global monitoring strategy. Nature 2015, 523, 403–406. [Google Scholar] [CrossRef] [PubMed]
- GOFC-GOLD. A Sourcebook of Methods and Procedures for Monitoring Essential Biodiversity Variables in Tropical Forests with Remote Sensing. In Report Version UNCBD COP-13; GOFC-GOLD Land Cover Project Office, Wageningen University: Wageningen, The Netherlands, 2017. [Google Scholar]
- Rocchini, D.; Boyd, D.S.; Féret, J.-B.; Foody, G.M.; He, K.S.; Lausch, A.; Nagendra, H.; Wegmann, M.; Pettorelli, N. Satellite remote sensing to monitor species diversity: Potential and pitfalls. Remote Sens. Ecol. Conserv. 2016, 2, 25–36. [Google Scholar] [CrossRef]
- Pettorelli, N.; Schulte to Bühne, H.; Tulloch, A.; Dubois, G.; Macinnis-Ng, C.; Queirós, A.M.; Keith, D.A.; Wegmann, M.; Schrodt, F.; Stellmes, M. Satellite remote sensing of ecosystem functions: Opportunities, challenges and way forward. Remote Sens. Ecol. Conserv. 2017. [Google Scholar] [CrossRef]
- Mora, B.; Szantoi, Z.; Heiden, U. Available online: http://elib.dlr.de/112264/1/BiodiversitySourcebook.pdf (accessed on 26 June 2017).
- Carlson, K.M.; Asner, G.P.; Hughes, R.F.; Ostertag, R.; Martin, R.E. Hyperspectral remote sensing of canopy biodiversity in Hawaiian lowland rainforests. Ecosystems 2007, 10, 536–549. [Google Scholar] [CrossRef]
- Ahumada, J.A.; Silva, C.E.; Gajapersad, K.; Hallam, C.; Hurtado, J.; Martin, E.; McWilliam, A.; Mugerwa, B.; O’Brien, T.; Rovero, F. Community structure and diversity of tropical forest mammals: Data from a global camera trap network. Philos. Trans. R. Soc. B-Biol. Sci. 2011, 366, 2703–2711. [Google Scholar] [CrossRef] [PubMed]
- Yao, W.; Krzystek, P.; Heurich, M. Tree species classification and estimation of stem volume and dbh based on single tree extraction by exploiting airborne full-waveform LiDAR data. Remote Sens. Environ. 2012, 123, 368–380. [Google Scholar] [CrossRef]
- Fisher, J.I.; Mustard, J.F.; Vadeboncoeur, M.A. Green leaf phenology at landsat resolution: Scaling from the field to the satellite. Remote Sens. Environ. 2006, 100, 265–279. [Google Scholar] [CrossRef]
- Atkinson, P.M.; Jeganathan, C.; Dash, J.; Atzberger, C. Inter-comparison of four models for smoothing satellite sensor time-series data to estimate vegetation phenology. Remote Sens. Environ. 2012, 123, 400–417. [Google Scholar] [CrossRef]
- Jay, S.; Potter, C.; Crabtree, R.; Genovese, V.; Weiss, D.J.; Kraft, M. Evaluation of modelled net primary production using modis and landsat satellite data fusion. Carbon Balance Manag. 2016, 11, 8. [Google Scholar] [CrossRef] [PubMed]
- Cleveland, C.C.; Taylor, P.; Chadwick, K.D.; Dahlin, K.; Doughty, C.E.; Malhi, Y.; Smith, W.K.; Sullivan, B.W.; Wieder, W.R.; Townsend, A.R. A comparison of plot-based satellite and earth system model estimates of tropical forest net primary production. Glob. Biogeochem. Cycles 2015, 29, 626–644. [Google Scholar] [CrossRef]
- Smith, M.-L.; Ollinger, S.V.; Martin, M.E.; Aber, J.D.; Hallett, R.A.; Goodale, C.L. Direct estimation of aboveground forest productivity through hyperspectral remote sensing of canopy nitrogen. Ecol. Appl. 2002, 12, 1286–1302. [Google Scholar] [CrossRef]
- Verma, M.; Friedl, M.A.; Law, B.; Bonal, D.; Kiely, G.; Black, T.; Wohlfahrt, G.; Moors, E.J.; Montagnani, L.; Marcolla, B. Improving the performance of remote sensing models for capturing intra-and inter-annual variations in daily gpp: An analysis using global fluxnet tower data. Agric. For. Meteorol. 2015, 214, 416–429. [Google Scholar] [CrossRef]
- Flores, B.M.; Piedade, M.-T.F.; Nelson, B.W. Fire disturbance in amazonian blackwater floodplain forests. Plant Ecol. Divers. 2014, 7, 319–327. [Google Scholar] [CrossRef]
- Jin, S.; Sader, S.A. Modis time-series imagery for forest disturbance detection and quantification of patch size effects. Remote Sens. Environ. 2005, 99, 462–470. [Google Scholar] [CrossRef]
- Ngoprasert, D.; Lynam, A.J.; Gale, G.A. Human disturbance affects habitat use and behaviour of asiatic leopard panthera pardus in kaeng krachan national park, Thailand. Oryx 2007, 41, 343–351. [Google Scholar] [CrossRef]
- Hansen, M.C.; Potapov, P.V.; Goetz, S.J.; Turubanova, S.; Tyukavina, A.; Krylov, A.; Kommareddy, A.; Egorov, A. Mapping tree height distributions in sub-saharan africa using landsat 7 and 8 data. Remote Sens. Environ. 2016, 185, 221–232. [Google Scholar] [CrossRef]
- Tuanmu, M.N.; Jetz, W. A global, remote sensing-based characterization of terrestrial habitat heterogeneity for biodiversity and ecosystem modelling. Glob. Ecol. Biogeogr. 2015, 24, 1329–1339. [Google Scholar] [CrossRef]
- Simonson, W.D.; Allen, H.D.; Coomes, D.A. Applications of airborne LiDAR for the assessment of animal species diversity. Methods Ecol. Evol. 2014, 5, 719–729. [Google Scholar] [CrossRef]
- Betbeder, J.; Hubert-Moy, L.; Burel, F.; Corgne, S.; Baudry, J. Assessing ecological habitat structure from local to landscape scales using synthetic aperture radar. Ecol. Indic. 2015, 52, 545–557. [Google Scholar] [CrossRef] [Green Version]
- Leblanc, S.G.; Fournier, R.A. Measurement of forest structure with hemispherical photography. In Hemispherical Photography in Forest Science: Theory, Methods, Applications; Springer: Berlin, Germany, 2017; pp. 53–83. [Google Scholar]
- Lehner, B.; Döll, P. Development and validation of a global database of lakes, reservoirs and wetlands. J. Hydrol. 2004, 296, 1–22. [Google Scholar] [CrossRef]
- Bruford, M.W.; Davies, N.; Dulloo, M.E.; Faith, D.P.; Walters, M. Monitoring changes in genetic diversity. In The GEO Handbook on Biodiversity Observation Networks; Walters, M., Scholes, R.J., Eds.; Springer International Publishing: Berlin, Germany, 2017; pp. 107–128. [Google Scholar]
- Vihervaara, P.; Auvinen, A.-P.; Mononen, L.; Törmä, M.; Ahlroth, P.; Anttila, S.; Böttcher, K.; Forsius, M.; Heino, J.; Heliölä, J. How essential biodiversity variables and remote sensing can help national biodiversity monitoring. Glob. Ecol. Conserv. 2017, 10, 43–59. [Google Scholar] [CrossRef]
- Obrist, M.K.; Pavan, G.; Sueur, J.; Riede, K.; Llusia, D.; Márquez, R. Bioacoustics approaches in biodiversity inventories. Abc Taxa 2010, 8, 68–99. [Google Scholar]
- Pereira, H.M.; Belnap, J.; Böhm, M.; Brummitt, N.; Garcia-Moreno, J.; Gregory, R.; Martin, L.; Peng, C.; Proença, V.; Schmeller, D.; et al. Monitoring essential biodiversity variables at the species level. In The GEO Handbook on Biodiversity Observation Networks; Walters, M., Scholes, R.J., Eds.; Springer International Publishing: Berlin, Germany, 2017; pp. 79–105. [Google Scholar]
- Rocchini, D.; Foody, G.M.; Nagendra, H.; Ricotta, C.; Anand, M.; He, K.S.; Amici, V.; Kleinschmit, B.; Förster, M.; Schmidtlein, S. Uncertainty in ecosystem mapping by remote sensing. Comput. Geosci. 2013, 50, 128–135. [Google Scholar] [CrossRef]
- Wrege, P.H.; Rowland, E.D.; Keen, S.; Shiu, Y. Acoustic monitoring for conservation in tropical forests: Examples from forest elephants. Methods Ecol. Evol. 2017. [Google Scholar] [CrossRef]
- Bucklin, A.; Lindeque, P.K.; Rodriguez-Ezpeleta, N.; Albaina, A.; Lehtiniemi, M. Metabarcoding of marine zooplankton: Prospects, progress and pitfalls. J. Plankton Res. 2016, 38, 393–400. [Google Scholar] [CrossRef]
- Paganini, M.; Leidner, A.K.; Geller, G.; Turner, W.; Wegmann, M. The role of space agencies in remotely sensed essential biodiversity variables. Remote Sens. Ecol. Conserv. 2016, 2, 132–140. [Google Scholar] [CrossRef]
- Geller, G.N.; Halpin, P.N.; Helmuth, B.; Hestir, E.L.; Skidmore, A.; Abrams, M.J.; Aguirre, N.; Blair, M.; Botha, E.; Colloff, M.; et al. Remote sensing for biodiversity. In The GEO Handbook on Biodiversity Observation Networks; Walters, M., Scholes, R.J., Eds.; Springer International Publishing: Berlin, Germany, 2017; pp. 187–210. [Google Scholar]
- Creer, S.; Deiner, K.; Frey, S.; Porazinska, D.; Taberlet, P.; Thomas, W.K.; Potter, C.; Bik, H.M. The ecologist’s field guide to sequence-based identification of biodiversity. Methods Ecol. Evol. 2016, 7, 1008–1018. [Google Scholar] [CrossRef]
- Jahn, O.; Ganchev, T.D.; Marques, M.I.; Schuchmann, K.-L. Automated sound recognition provides insights into the behavioral ecology of a tropical bird. PLoS ONE 2017, 12, e0169041. [Google Scholar] [CrossRef] [PubMed]
- Jeliazkov, A.; Bas, Y.; Kerbiriou, C.; Julien, J.-F.; Penone, C.; Le Viol, I. Large-scale semi-automated acoustic monitoring allows to detect temporal decline of bush-crickets. Glob. Ecol. Conserv. 2016, 6, 208–218. [Google Scholar] [CrossRef]
- O’Brien, T. On the use of automated cameras to estimate species richness for large-and medium-sized rainforest mammals. Anim. Conserv. 2008, 11, 179–181. [Google Scholar] [CrossRef]
- Ivanova, N.V.; Dewaard, J.R.; Hebert, P.D. An inexpensive, automation-friendly protocol for recovering high-quality DNA. Mol. Ecol. Resour. 2006, 6, 998–1002. [Google Scholar] [CrossRef]
- Forrester, T.; O’Brien, T.; Fegraus, E.; Jansen, P.A.; Palmer, J.; Kays, R.; Ahumada, J.; Stern, B.; McShea, W. An open standard for camera trap data. Biodivers. Data J. 2016. [Google Scholar] [CrossRef] [PubMed]
- Roch, M.A.; Batchelor, H.; Baumann-Pickering, S.; Berchok, C.L.; Cholewiak, D.; Fujioka, E.; Garland, E.C.; Herbert, S.; Hildebrand, J.A.; Oleson, E.M. Management of acoustic metadata for bioacoustics. Ecol. Inform. 2016, 31, 122–136. [Google Scholar] [CrossRef] [Green Version]
- Ratnasingham, S.; Hebert, P.D. Bold: The barcode of life data system (http://www.barcodinglife.org). Mol. Ecol. Resour. 2007, 7, 355–364. [Google Scholar] [CrossRef] [PubMed]
- Aide, T.M.; Corrada-Bravo, C.; Campos-Cerqueira, M.; Milan, C.; Vega, G.; Alvarez, R. Real-time bioacoustics monitoring and automated species identification. PeerJ 2013, 1, e103. [Google Scholar] [CrossRef] [PubMed]
- Bohmann, K.; Evans, A.; Gilbert, M.T.P.; Carvalho, G.R.; Creer, S.; Knapp, M.; Douglas, W.Y.; de Bruyn, M. Environmental DNA for wildlife biology and biodiversity monitoring. Trends Ecol. Evol. 2014, 29, 358–367. [Google Scholar] [CrossRef] [PubMed]







| Compositional Biodiversity | Functional Biodiversity | Structural Biodiversity | ||
|---|---|---|---|---|
| In situ | Established field survey |  |  |  |
| In situ activity sensors |  |  |  | |
| DNA Barcoding |  |  |  | |
| Citizen Science |  |  |  | |
| Remote sensing | Coarse to medium spatial resolution |  |  |  |
| High to very high spatial resolution |  |  |  | |
| Hyperspectral |  |  |  | |
| SAR |  |  |  | |
| Airborne LiDAR |  |  |  | |
| Terrestrial Laser Scanner |  |  |  | |
| EBV Classes | EBV Candidates | Aichi Target [10] | Remote Sensing | Emerging Opportunities |
|---|---|---|---|---|
| Species Populations | Species distribution | 4–12, 14, 15 | High to very high SR [49], Hyperspectral [89] | In situ activity sensors [90] DNA barcoding [36] |
| Population abundance | 4–12, 14, 15 | High to very high SR [49], Hyperspectral [89] | In situ activity sensors [90] | |
| Population structure by age/size class | 4–12, 14, 15 | LiDAR [91] | ||
| Species Traits | Phenology | 10, 15 | High to very high SR [92], coarse to medium SR [93], hyperspectral [72] | |
| Migratory behaviour | 5, 6, 10, 11, 12 | In situ activity sensors [35] | ||
| Community Composition | Taxonomic diversity | 8, 10, 12, 14 | Hyperspectral [29] | In situ activity sensors DNA barcoding [36] |
| Ecosystem Function | Net primary productivity | 5, 8, 14 | High to very high SR [94], coarse to medium SR [95], hyperspectral [96] | In situ activity sensors [97] |
| Nutrient retention | 5, 8, 14 | Hyperspectral [96] | ||
| Disturbance regime | 5, 7, 9, 10, 11, 14, 15 | High to very high SR [98], coarse to medium SR [99] | In situ activity sensors [100] | |
| Ecosystem Structure | Habitat structure | 5, 11, 14, 15 | High to very high SR [101], coarse to medium SR [102], LiDAR [103], SAR [104] | In situ activity sensors [105] |
| Ecosystem extent and fragmentation | 5, 7, 10, 14, 15 | coarse to medium SR [106] |
| EBV Criteria Components | Remote Sensing | In Situ | DNA Barcoding |
|---|---|---|---|
| Spatial extent | Global [57] | Global with gaps. Example: TEAM network (http://www.teamnetwork.org/), http://bio.acousti.ca Bioacoustics [60] | Local/regional [107] |
| Spatial resolution | Optical satellite: coarse spatial resolution 250–1200 m (e.g., MODIS), Medium to high spatial resolution: 5–30 m (e.g., Landsat, sentinel 2, RapidEye), Very high spatial resolution (e.g., Ikonos, GeoEye): 0.5–4 m. Airborne Hyperspectral: 1–2 m (according to flight height). Active remote sensing (radar): 1–100 m [108]. Upcoming: GEDI (satellite LiDAR): 25 m footprint, EnMAP (satellite hyperspectral): 250 narrow bands [88] | Field based. Example: TEAM has 23 tropical forest sites (120–200 km2 resolution) [15] | Requires physical sampling [107] |
| Periodicity | Continuous long term time-series data, with high revisit-time period for high-resolution data (e.g., Landsat: every 16 days, Sentinel 2: every 10 days, RapidEye: Daily ) [108] | From real-time to different times of the day and seasons [109] | No clear understanding [107] |
| Taxonomic coverage | Multiple taxa can be covered [110] | Multiple taxa can be covered [110] | Multiple taxa can be covered [110] |
| uncertainty | Imperfect detections, data uncertainties, model uncertainties [111] | Measurement error, detection algorithms [112], spatial mismatches [69] | Reference datasets [15], variation in primer use, amplification steps and sequencing platforms [113] |
| Operational definition | Several demonstrations are made to derive EBVs [85,87,114,115] | The technology has been identified as candidate [57] | The technology has been identified as candidate [19,116] |
| Documentation | Documentations is available [85,87,115] | Lack of documentation and established protocols | Lack of documentation and established protocols |
| Abstraction | Few to several steps involved in derivation of products [85,87] | Few steps involved in derivation of products [110] | Several steps in derivation of products [116] |
| Measurement and sampling schema | Sampling and measuring strategies are often well defined [87] | Limited sampling and measuring strategies are available [14]. Camera traps: www.teamnetwork.org/protocols | Few sampling and measuring strategies are available [116] (www.biocodecommons.org/, www.gensc.org/) |
| Automatisation | Automation of data acquisition and processing is possible [115] | Automation of data acquisition, processing, and management are possible. Example: automated and semi-automated sound recognition [117,118], automated camera traps and image recognition [119] | Automated DNA extraction is possible [120] |
| Interoperability | Global standards and protocols exist for harmonised data and metadata formats (e.g., http://docs.opengeospatial.org/is/10-157r4/10-157r4.html) | Camera traps: individual initiatives exist [121], Bioacoustics: metadata standards are proposed [122] | Data standards are defined [123] |
| Data availability | Data available for multiple EBVs [85,87]. (e.g. https://scihub.copernicus.eu/dhus/#/home, https://gcmd.nasa.gov/, https://boninabox.geobon.org/) | Data mobilisation opportunities exist www.TEAMNetwork.org, http://bio.acousti.ca/, https://boninabox.geobon.org/, https://www.movebank.org/ | Data mobilisation opportunities exist http://www.barcodinglife.org, https://0-www-ncbi-nlm-nih-gov.brum.beds.ac.uk/, https://boninabox.geobon.org/ |
| Temporal sustainability | Data have been available from satellite agencies for 40 years now (e.g., Landsat) and is secured until the end of the 2020’s [108,114] | Data availability and methods are evolving [124] | Data availability and methods are evolving [18] |
| Baseline | Historical satellite datasets are available: e.g., Landsat program (since 1972) [108] | Baselines can be made from past field inventories [112] | Ancient DNA (e.g., from museum collections) [125], https://0-www-ncbi-nlm-nih-gov.brum.beds.ac.uk/ |
| Relevancy | Relevance for multiple EBVs has been demonstrated [87,108] | Relevance for multiple EBVs has been demonstrated [110,112] | Relevance for multiple EBVs has been demonstrated [107,110] |
| Consensus | Large consensus exists [85] | Consensus underway [110] | Consensus underway [107] |
| Scalability | Robust to scalability (e.g., diversity indices) [86] | Robust to scalability (e.g., Wildlife Picture Index) [75] | Robust to scalability using statistical models (e.g., species distribution models) [19] |
| Institutional support | Several institutions are contributing. Example: GEO BON (http://geobon.org/essential-biodiversity-variables/monitoring/), GOFC-GOLD: (http://www.gofcgold.wur.nl) | Several institutions are contributing. Example: GEO BON [14], Map Of Life (https://mol.org/), Move bank (https://www.movebank.org/) | GEO BON (http://geobon.org/essential-biodiversity-variables/monitoring/), GOFC-GOLD: (http://www.gofcgold.wur.nl) |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mulatu, K.A.; Mora, B.; Kooistra, L.; Herold, M. Biodiversity Monitoring in Changing Tropical Forests: A Review of Approaches and New Opportunities. Remote Sens. 2017, 9, 1059. https://0-doi-org.brum.beds.ac.uk/10.3390/rs9101059
Mulatu KA, Mora B, Kooistra L, Herold M. Biodiversity Monitoring in Changing Tropical Forests: A Review of Approaches and New Opportunities. Remote Sensing. 2017; 9(10):1059. https://0-doi-org.brum.beds.ac.uk/10.3390/rs9101059
Chicago/Turabian StyleMulatu, Kalkidan Ayele, Brice Mora, Lammert Kooistra, and Martin Herold. 2017. "Biodiversity Monitoring in Changing Tropical Forests: A Review of Approaches and New Opportunities" Remote Sensing 9, no. 10: 1059. https://0-doi-org.brum.beds.ac.uk/10.3390/rs9101059