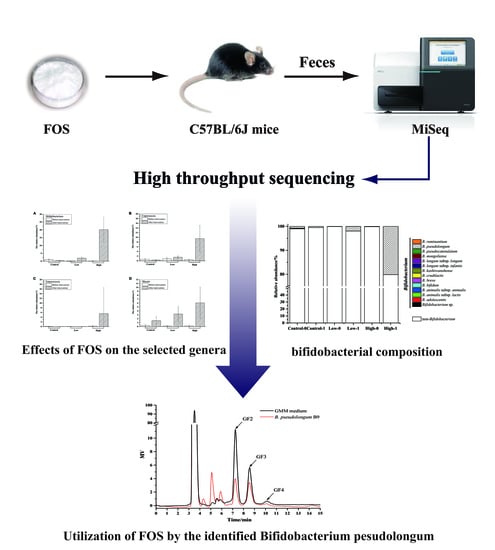
Effects of Different Doses of Fructooligosaccharides (FOS) on the Composition of Mice Fecal Microbiota, Especially the Bifidobacterium Composition
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals and Reagents
2.2. Animal Experiment Design and Sample Collection
2.3. Genomic DNA Extraction and PCR Amplification of the V3–V4 Region and the groEL Gene
2.4. Quantification and Sequencing
2.5. Bioinformatic Analysis
2.6. Isolation and Identification of Bifidobacteria from Mice Feces
2.7. Growth of Bifidobacteria on FOS
2.8. Statistical Analysis
3. Results
3.1. Body Weight and Dietary Intake
3.2. Short-Chain Fatty Acids (SCFAs) Determination in Feces of Mice
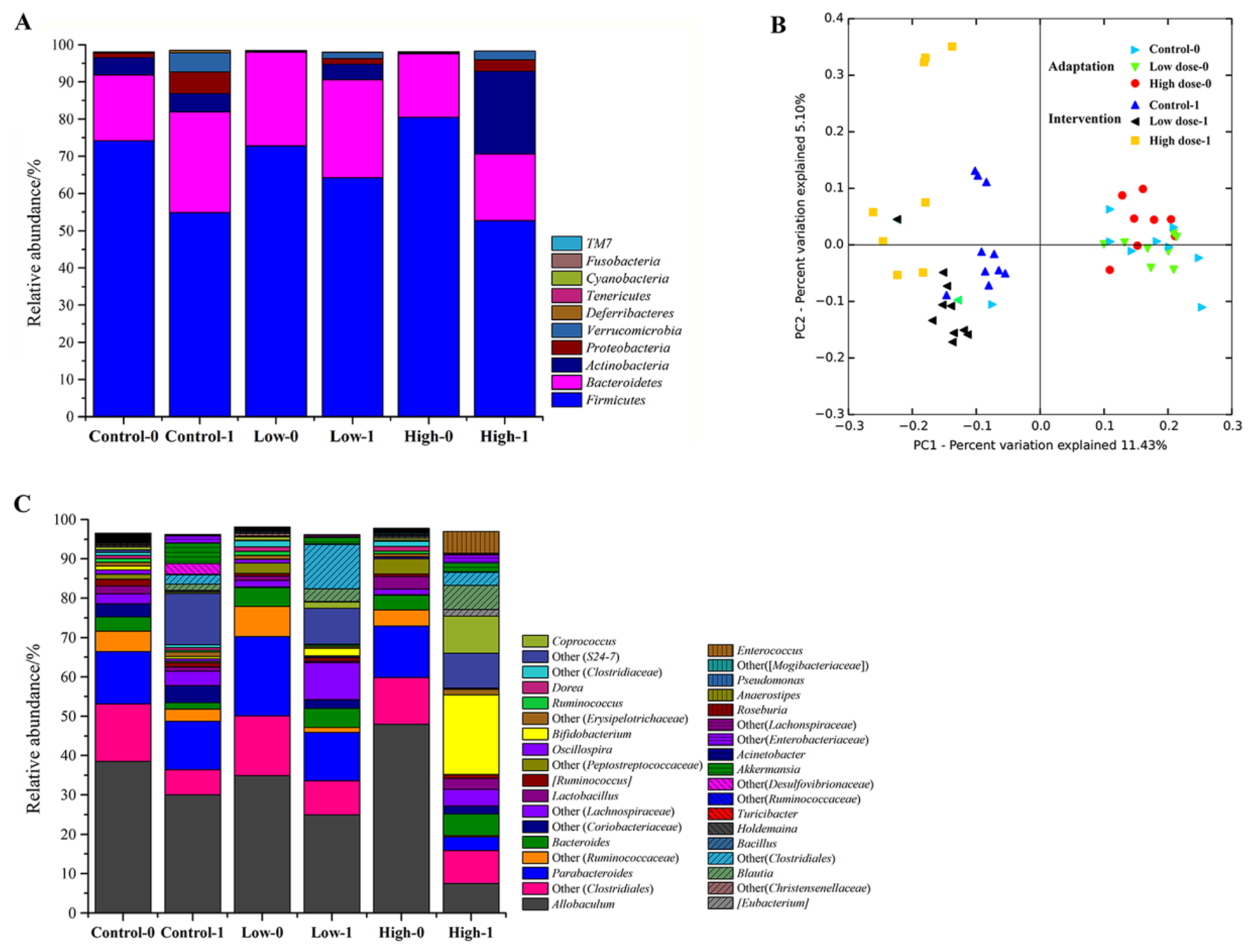
3.3. The Effects of FOS on the Fecal Microbiota of Mice
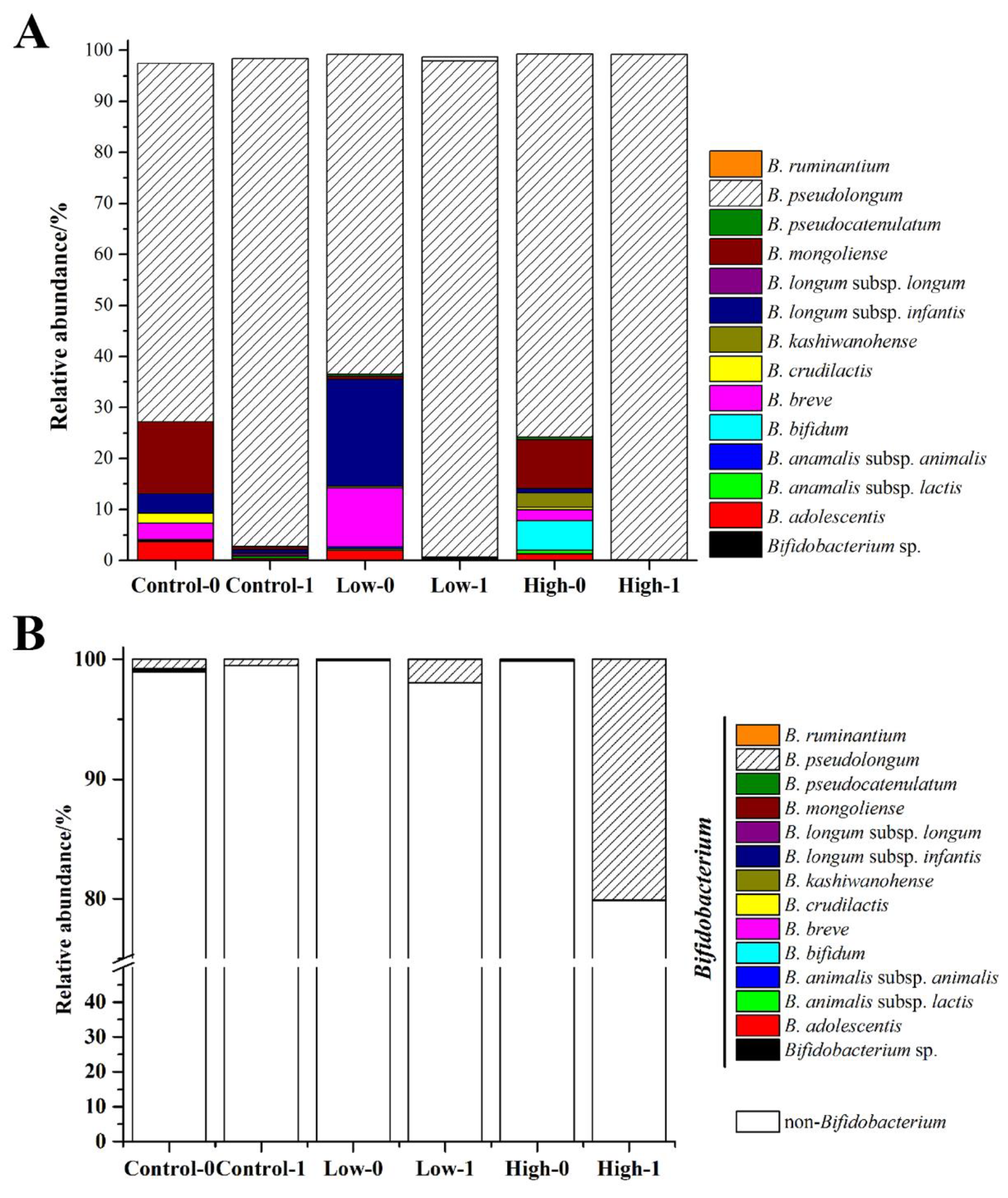
3.4. The Effects of FOS on the Composition of Bifidobacterium in Feces
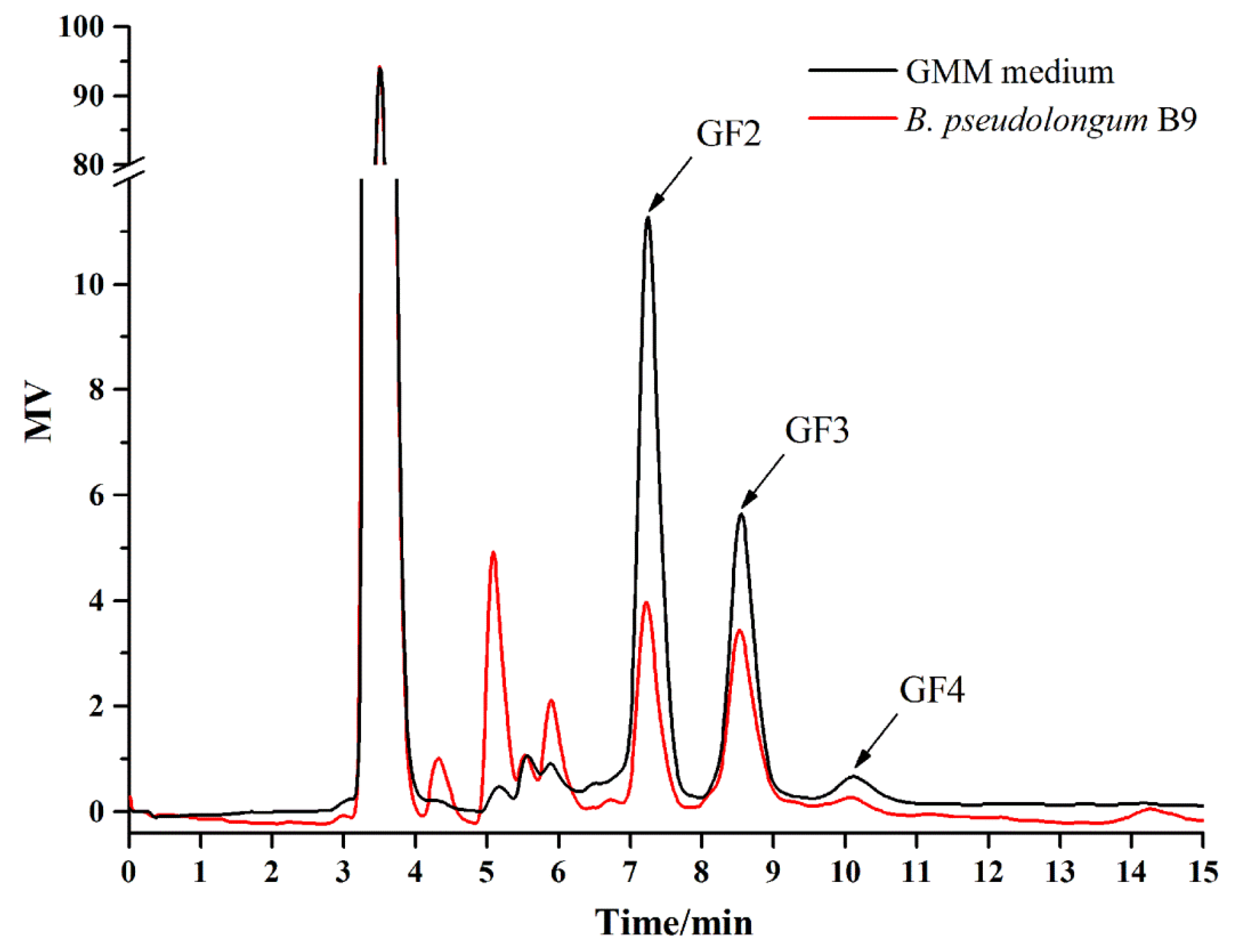
3.5. Isolation of B. pseudolongum from Mice Feces and Characterization of Its Growth on FOS
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Backhed, F.; Ley, R.E.; Sonnenburg, J.L.; Peterson, D.A.; Gordon, J.I. Host-bacterial mutualism in the human intestine. Science 2005, 307, 1915–1920. [Google Scholar] [CrossRef] [PubMed]
- Sekirov, I.; Russell, S.L.; Antunes, L.C.M.; Finlay, B.B. Gut microbiota in health and disease. Physiol. Rev. 2010, 90, 859–904. [Google Scholar] [CrossRef] [PubMed]
- Collins, M.D.; Gibson, G.R. Probiotics, prebiotics, and synbiotics: Approaches for modulating the microbial ecology of the gut. Am. J. Clin. Nutr. 1999, 69, 1052s–1057s. [Google Scholar] [CrossRef] [PubMed]
- Picard, C.; Fioramonti, J.; Francois, A.; Robinson, T.; Neant, F.; Matuchansky, C. Review article: Bifidobacteria as probiotic agents—Physiological effects and clinical benefits. Aliment. Pharm. Ther. 2005, 22, 495–512. [Google Scholar] [CrossRef] [PubMed]
- Turnbaugh, P.J.; Ley, R.E.; Mahowald, M.A.; Magrini, V.; Mardis, E.R.; Gordon, J.I. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 2006, 444, 1027–1031. [Google Scholar] [CrossRef] [PubMed]
- LeBlanc, J.G.; Milani, C.; de Giori, G.S.; Sesma, F.; van Sinderen, D.; Ventura, M. Bacteria as vitamin suppliers to their host: A gut microbiota perspective. Curr. Opin. Biotechnol. 2013, 24, 160–168. [Google Scholar] [CrossRef] [PubMed]
- Kleerebezem, M.; Vaughan, E.E. Probiotic and gut Lactobacilli and Bifidobacteria: Molecular approaches to study diversity and activity. Annu. Rev. Microbiol. 2009, 63, 269–290. [Google Scholar] [CrossRef] [PubMed]
- Gibson, G.R.; Hutkins, R.; Sanders, M.E.; Prescott, S.L.; Reimer, R.A.; Salminen, S.J.; Scott, K.; Stanton, C.; Swanson, K.S.; Cani, P.D.; et al. The international scientific association for probiotics and prebiotics (ISAPP) consensus statement on the definition and scope of prebiotics. Nat. Rev. Gastroenterol. Hepatol. 2017, 14, 491–502. [Google Scholar] [CrossRef] [PubMed]
- Gibson, G.R.; Roberfroid, M.B. Dietary modulation of the human colonic microbiota—Introducing the concept of prebiotics. J. Nutr. 1995, 125, 1401–1412. [Google Scholar] [CrossRef] [PubMed]
- Roberfroid, M.B. Prebiotics: Preferential substrates for specific germs? Am. J. Clin. Nutr. 2001, 73, 406s–409s. [Google Scholar] [CrossRef] [PubMed]
- Gibson, G.R.; Beatty, E.R.; Wang, X.; Cummings, J.H. Selective stimulation of Bifidobacteria in the human colon by oligofructose and inulin. Gastroenterology 1995, 108, 975–982. [Google Scholar] [CrossRef]
- Mao, B.Y.; Li, D.Y.; Zhao, J.X.; Liu, X.M.; Gu, Z.N.; Chen, Y.Q.; Zhang, H.; Chen, W. In vitro fermentation of fructooligosaccharides with human gut bacteria. Food Funct. 2015, 6, 947–954. [Google Scholar] [CrossRef] [PubMed]
- Vigsnaes, L.K.; Holck, J.; Meyer, A.S.; Licht, T.R. In vitro fermentation of sugar beet arabino-oligosaccharides by fecal microbiota obtained from patients with ulcerative colitis to selectively stimulate the growth of Bifidobacterium spp. and Lactobacillus spp. Appl. Environ. Microbiol. 2011, 77, 8336–8344. [Google Scholar] [CrossRef] [PubMed]
- Junick, J.; Blaut, M. Quantification of human fecal Bifidobacterium species by use of quantitative real-time PCR analysis targeting the groEL gene. Appl. Environ. Microbiol. 2012, 78, 2613–2622. [Google Scholar] [CrossRef] [PubMed]
- Hu, L.J.; Lu, W.W.; Wang, L.L.; Pan, M.L.; Zhang, H.; Zhao, J.X.; Chen, W. Assessment of Bifidobacterium species using groEL Gene on the basis of Illumina MiSeq high-throughput sequencing. Genes 2017, 8, 336. [Google Scholar] [CrossRef] [PubMed]
- Campbell, J.M.; Fahey, G.C.; Wolf, B.W. Selected indigestible oligosaccharides affect large bowel mass, cecal and fecal short-chain fatty acids, pH and microflora in rats. J. Nutr. 1997, 127, 130–136. [Google Scholar] [CrossRef] [PubMed]
- Mao, B.Y.; Li, D.Y.; Zhao, J.X.; Liu, X.M.; Gu, Z.N.; Chen, Y.Q.; Zhang, H.; Chen, W. Metagenomic insights into the effects of fructo-oligosaccharides (FOS) on the composition of fecal microbiota in mice. J. Agric. Food Chem. 2015, 63, 856–863. [Google Scholar] [CrossRef] [PubMed]
- Mao, B.Y.; Li, D.Y.; Ai, C.Q.; Zhao, J.X.; Zhang, H.; Chen, W. Lactulose differently modulates the composition of luminal and mucosal microbiota in C57BL/6J mice. J. Agric. Food Chem. 2016, 64, 6240–6247. [Google Scholar] [CrossRef] [PubMed]
- Hill, J.E.; Penny, S.L.; Crowell, K.G.; Goh, S.H.; Hemmingsen, S.M. cpnDB: A chaperonin sequence database. Genome Res. 2004, 14, 1669–1675. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef] [PubMed]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Pena, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Caporaso, J.G.; Bittinger, K.; Bushman, F.D.; DeSantis, T.Z.; Andersen, G.L.; Knight, R. PyNAST: A flexible tool for aligning sequences to a template alignment. Bioinformatics 2010, 26, 266–267. [Google Scholar] [CrossRef] [PubMed]
- Lozupone, C.; Knight, R. UniFrac: A new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 2005, 71, 8228–8235. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Wang, G.; Siegel, P.; He, C.; Wang, H.; Zhao, W.; Zhai, Z.; Tian, F.; Zhao, J.; Zhang, H.; et al. Quantitative genetic background of the host influences gut microbiomes in chickens. Sci. Rep. 2013, 3, 1163. [Google Scholar] [CrossRef] [PubMed]
- Coussement, P.A. Inulin and oligofructose: Safe intakes and legal status. J. Nutr. 1999, 129, 1412S–1417S. [Google Scholar] [CrossRef] [PubMed]
- Kawaguchi, M.; Tashiro, Y.; Adachi, T.; Tamura, Z. Changes in intestinal condition, fecal microflora and composition of rectal gas after administration of fructooligosaccharide and lactulose at different doses. Biosci. Microflora 1993, 12, 57–67. [Google Scholar] [CrossRef]
- David, L.A.; Maurice, C.F.; Carmody, R.N.; Gootenberg, D.B.; Button, J.E.; Wolfe, B.E.; Ling, A.V.; Devlin, A.S.; Varma, Y.; Fischbach, M.A.; et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature 2014, 505, 559–563. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shen, J.A.; Zhang, B.R.; Wei, H.; Che, C.Y.; Ding, D.Z.; Hua, X.G.; Bucheli, P.; Wang, L.H.; Li, Y.X.; Pang, X.Y.; et al. Assessment of the modulating effects of fructo-oligosaccharides on fecal microbiota using human flora-associated piglets. Arch. Microbiol. 2010, 192, 959–968. [Google Scholar] [CrossRef] [PubMed]
- Mikkelsen, L.L.; Jensen, B.B. Effect of fructo-oligosaccharides and transgalacto-oligosaccharides on microbial populations and microbial activity in the gastrointestinal tract of piglets post-weaning. Anim. Feed Sci. Technol. 2004, 117, 107–119. [Google Scholar] [CrossRef]
- Shadid, R.; Haarman, M.; Knol, J.; Theis, W.; Beermann, C.; Rjosk-Dendorfer, D.; Schendel, D.J.; Koletzko, B.V.; Krauss-Etschmann, S. Effects of galactooligosaccharide and long-chain fructooligosaccharide supplementation during pregnancy on maternal and neonatal microbiota and immunity—A randomized, double-blind, placebo-controlled study. Am. J. Clin. Nutr. 2007, 86, 1426–1437. [Google Scholar] [CrossRef] [PubMed]
- Duranti, S.; Milani, C.; Lugli, G.A.; Turroni, F.; Mancabelli, L.; Sanchez, B.; Ferrario, C.; Viappiani, A.; Mangifesta, M.; Mancino, W.; et al. Insights from genomes of representatives of the human gut commensal Bifidobacterium bifidum. Environ. Microbiol. 2015, 17, 2515–2531. [Google Scholar] [CrossRef] [PubMed]
- Hopkins, M.J.; Cummings, J.H.; Macfarlane, G.T. Inter-species differences in maximum specific growth rates and cell yields of bifidobacteria cultured on oligosaccharides and other simple carbohydrate sources. J. Appl. Microbiol. 1998, 85, 381–386. [Google Scholar] [CrossRef]
- Hidaka, T.; Eida, T.; Takizawa, T.; Tokunaga, T.; Tashiro, Y. Effects of fructooligosaccharides on intestinal flora and human health. Biosci. Microflora 1986, 5, 37–50. [Google Scholar] [CrossRef]
- Scott, K.P.; Martin, J.C.; Duncan, S.H.; Flint, H.J. Prebiotic stimulation of human colonic butyrate-producing bacteria and bifidobacteria, in vitro. FEMS Microbiol. Ecol. 2014, 87, 30–40. [Google Scholar] [CrossRef] [PubMed]
- Linke, C.M.; Woodiga, S.A.; Meyers, D.J.; Buckwalter, C.M.; Salhi, H.E.; King, S.J. The ABC Transporter Encoded at the Pneumococcal fructooligosaccharide utilization locus determines the ability to utilize long- and short-chain fructooligosaccharides. J. Bacteriol. 2013, 195, 1031–1041. [Google Scholar] [CrossRef] [PubMed]
- Eren, A.M.; Sogin, M.L.; Morrison, H.G.; Vineis, J.H.; Fisher, J.C.; Newton, R.J.; McLellan, S.L. A single genus in the gut microbiome reflects host preference and specificity. ISME J. 2015, 9, 90–100. [Google Scholar] [CrossRef] [PubMed]
- Biddle, A.; Stewart, L.; Blanchard, J.; Leschine, S. Untangling the genetic basis of fibrolytic specialization by Lachnospiraceae and Ruminococcaceae in diverse gut communities. Diversity 2013, 5, 627–640. [Google Scholar] [CrossRef]
- Flint, H.J.; Bayer, E.A.; Rincon, M.T.; Lamed, R.; White, B.A. Polysaccharide utilization by gut bacteria: Potential for new insights from genomic analysis. Nat. Rev. Microbiol. 2008, 6, 121–131. [Google Scholar] [CrossRef] [PubMed]



| Group | Glu | HDL-C | LDL-C | HDL-C/LDL-C | TC | TG |
|---|---|---|---|---|---|---|
| Control | 4.61 ± 0.77 | 3.06 ± 0.75 | 0.38 ± 0.09 | 8.15 ± 0.88 | 3.45 ± 0.91 | 1.14 ± 0.28 |
| Low-dose | 7.73 ± 2.14 | 3.28 ± 0.24 | 0.40 ± 0.04 | 8.26 ± 0.92 | 3.72 ± 0.31 | 1.03 ± 0.47 |
| High-dose | 5.77 ± 1.95 | 2.47 ± 0.25 | 0.35 ± 0.13 | 7.81 ± 2.54 | 2.83 ± 0.38 | 0.70 ± 0.18 |
| p1 | NS | NS | NS | NS | NS | NS |
| SCFAs, μmol/g 1 | Control | Low-Dose | High-Dose | p2 |
|---|---|---|---|---|
| Acetic acid | 36.46 ± 10.83 | 25.61 ± 15.20 | 45.24 ± 20.02 | NS |
| Propionic acid | 24.37 ± 7.90 | 17.55 ± 6.92 | 31.25 ± 9.44 | NS |
| Butyric acid | 20.07 ± 7.46 | 7.43 ± 5.67 | 13.78 ± 12.24 | NS |
| Isobutyric acid | 19.74 ± 6.17 | 14.45 ± 7.08 | 31.08 ± 15.81 | NS |
| Isovaleric acid | 14.53 ± 5.36 | 10.45 ± 3.93 | 18.58 ± 5.17 | NS |
| Valeric acid | 10.25 ± 3.60 | 7.17 ± 2.90 | 16.52 ± 9.94 | NS |
| Total | 125.41 ± 41.23 bc | 80.64 ± 28.38 bc | 156.44 ± 51.20 a | <0.05 |
| Group | Control | Low-dose | High-dose | ||||||
|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | p | 0 | 1 | p | 0 | 1 | p | |
| Shannon Index (SI) | 5.29 ± 1.49 | 5.37 ± 0.35 | >0.05 | 4.99 ± 0.64 | 5.60 ± 0.51 | <0.05 | 4.32 ± 0.68 | 5.01 ± 0.75 | >0.05 |
| No. | Strain | Most Positive Match | Ident (%) | GenBank |
|---|---|---|---|---|
| 1 | B9 | Bifidobacterium pseudolongum PV8-2 | 99% | MG820037 |
| 2 | B11 | Bifidobacterium pseudolongum strain UMB-MBP-01 | 99% | MG820038 |
| 3 | B24 | Bifidobacterium pseudolongum strain UMB-MBP-01 | 99% | MG820039 |
| 4 | B29 | Bifidobacterium pseudolongum strain UMB-MBP-01 | 99% | MG820040 |
| 5 | B72 | Bifidobacterium pseudolongum strain UMB-MBP-01 | 99% | MG820041 |
| 6 | B129 | Bifidobacterium pseudolongum PV8-2 | 99% | MG820042 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mao, B.; Gu, J.; Li, D.; Cui, S.; Zhao, J.; Zhang, H.; Chen, W. Effects of Different Doses of Fructooligosaccharides (FOS) on the Composition of Mice Fecal Microbiota, Especially the Bifidobacterium Composition. Nutrients 2018, 10, 1105. https://0-doi-org.brum.beds.ac.uk/10.3390/nu10081105
Mao B, Gu J, Li D, Cui S, Zhao J, Zhang H, Chen W. Effects of Different Doses of Fructooligosaccharides (FOS) on the Composition of Mice Fecal Microbiota, Especially the Bifidobacterium Composition. Nutrients. 2018; 10(8):1105. https://0-doi-org.brum.beds.ac.uk/10.3390/nu10081105
Chicago/Turabian StyleMao, Bingyong, Jiayu Gu, Dongyao Li, Shumao Cui, Jianxin Zhao, Hao Zhang, and Wei Chen. 2018. "Effects of Different Doses of Fructooligosaccharides (FOS) on the Composition of Mice Fecal Microbiota, Especially the Bifidobacterium Composition" Nutrients 10, no. 8: 1105. https://0-doi-org.brum.beds.ac.uk/10.3390/nu10081105