Dysregulation of Neuronal Genes by Fetal-Neonatal Iron Deficiency Anemia Is Associated with Altered DNA Methylation in the Rat Hippocampus
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Hippocampal Dissection
2.3. Whole Genome Bisulfite Sequencing and Library Preparation
2.4. Identification of DMRs Using the Defiant Program
2.5. Bioinformatics
3. Results
3.1. Early-Life Iron Deficiency Induced Differential DNA Methylation in the Rat Hippocampus
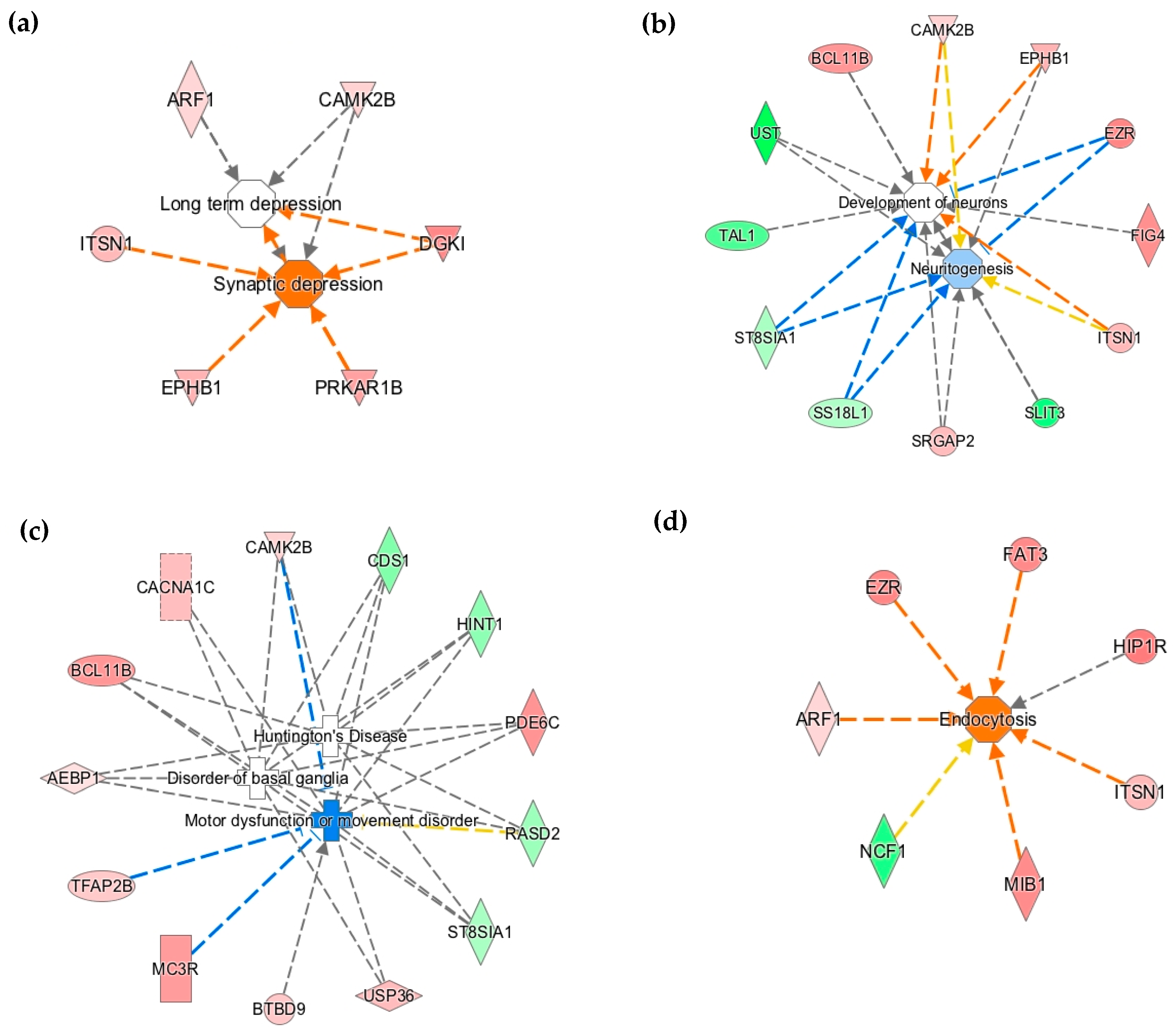
3.2. Early-Life Iron Deficiency Altered the Methylation Status of Genes Regulating Neuronal Development and Function
3.3. The Methylation Status of Genes Regulating Axonal Guidance Was Altered in the P15 ID Hippocampus
3.4. Differential DNA Methylation is a Potential Epigenetic Mechanism Contributing to Neural Gene Dysregulation in the P15 ID Hippocampus
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Lozoff, B.; Beard, J.; Connor, J.; Barbara, F.; Georgieff, M.; Schallert, T. Long-lasting neural and behavioral effects of iron deficiency in infancy. Nutr. Rev. 2006, 64, S34–S91. [Google Scholar] [CrossRef] [PubMed]
- Insel, B.J.; Schaefer, C.A.; McKeague, I.W.; Susser, E.S.; Brown, A.S. Maternal iron deficiency and the risk of schizophrenia in offspring. Arch. Gen. Psychiatry 2008, 65, 1136–1144. [Google Scholar] [CrossRef]
- Christian, P.; Murray-Kolb, L.E.; Khatry, S.K.; Katz, J.; Schaefer, B.A.; Cole, P.M.; Leclerq, S.C.; Tielsch, J.M. Prenatal micronutrient supplementation and intellectual and motor function in early school-aged children in Nepal. JAMA 2010, 304, 2716–2723. [Google Scholar] [CrossRef]
- Li, Q.; Yan, H.; Zeng, L.; Cheng, Y.; Liang, W.; Dang, S.; Wang, Q.; Tsuji, I. Effects of maternal multimicronutrient supplementation on the mental development of infants in rural western China: Follow-up evaluation of a double-blind, randomized, controlled trial. Pediatrics 2009, 123, e685–e692. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, M.K.; Muslimatun, S.; West, C.E.; Schultink, W.; Hautvast, J.G. Mental and psychomotor development in Indonesian infants of mothers supplemented with vitamin A in addition to iron during pregnancy. Br. J. Nutr. 2004, 91, 279–286. [Google Scholar] [CrossRef] [PubMed]
- Chang, S.; Zeng, L.; Brouwer, I.D.; Kok, F.J.; Yan, H. Effect of Iron Deficiency Anemia in Pregnancy on Child Mental Development in Rural China. Pediatrics 2013. [Google Scholar] [CrossRef]
- Siddappa, A.M.; Georgieff, M.K.; Wewerka, S.; Worwa, C.; Nelson, C.A.; Deregnier, R.A. Iron deficiency alters auditory recognition memory in newborn infants of diabetic mothers. Pediatr. Res. 2004, 55, 1034–1041. [Google Scholar] [CrossRef]
- Rao, R.; Georgieff, M.K. Iron in fetal and neonatal nutrition. Semin. Fetal Neonatal Med. 2007, 12, 54–63. [Google Scholar] [CrossRef] [Green Version]
- Rao, R.; Tkac, I.; Townsend, E.L.; Gruetter, R.; Georgieff, M.K. Perinatal iron deficiency alters the neurochemical profile of the developing rat hippocampus. J. Nutr. 2003, 133, 3215–3221. [Google Scholar] [PubMed]
- Avishai-Eliner, S.; Brunson, K.L.; Sandman, C.A.; Baram, T.Z. Stressed-out, or in (utero)? Trends Neurosci. 2002, 25, 518–524. [Google Scholar] [PubMed]
- Semple, B.D.; Blomgren, K.; Gimlin, K.; Ferriero, D.M.; Noble-Haeusslein, L.J. Brain development in rodents and humans: Identifying benchmarks of maturation and vulnerability to injury across species. Prog. Neurobiol. 2013, 106–107, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Petry, C.D.; Eaton, M.A.; Wobken, J.D.; Mills, M.M.; Johnson, D.E.; Georgieff, M.K. Iron deficiency of liver, heart, and brain in newborn infants of diabetic mothers. J. Pediatr. 1992, 121, 109–114. [Google Scholar] [CrossRef]
- Georgieff, M.K.; Petry, C.D.; Wobken, J.D.; Oyer, C.E. Liver and brain iron deficiency in newborn infants with bilateral renal agenesis (Potter’s syndrome). Pediatr. Pathol. Lab. Med. 1996, 16, 509–519. [Google Scholar] [CrossRef] [PubMed]
- Unger, E.L.; Hurst, A.R.; Georgieff, M.K.; Schallert, T.; Rao, R.; Connor, J.R.; Kaciroti, N.; Lozoff, B.; Felt, B. Behavior and monoamine deficits in prenatal and perinatal iron deficiency are not corrected by early postnatal moderate-iron or high-iron diets in rats. J. Nutr. 2012, 142, 2040–2049. [Google Scholar] [CrossRef]
- Schmidt, A.T.; Waldow, K.J.; Grove, W.M.; Salinas, J.A.; Georgieff, M.K. Dissociating the long-term effects of fetal/neonatal iron deficiency on three types of learning in the rat. Behav. Neurosci. 2007, 121, 475–482. [Google Scholar] [CrossRef] [PubMed]
- Felt, B.T.; Beard, J.L.; Schallert, T.; Shao, J.; Aldridge, J.W.; Connor, J.R.; Georgieff, M.K.; Lozoff, B. Persistent neurochemical and behavioral abnormalities in adulthood despite early iron supplementation for perinatal iron deficiency anemia in rats. Behav. Brain Res. 2006, 171, 261–270. [Google Scholar] [CrossRef] [Green Version]
- Brunette, K.E.; Tran, P.V.; Wobken, J.D.; Carlson, E.S.; Georgieff, M.K. Gestational and neonatal iron deficiency alters apical dendrite structure of CA1 pyramidal neurons in adult rat hippocampus. Dev. Neurosci. 2010, 32, 238–248. [Google Scholar] [CrossRef]
- Jorgenson, L.A.; Wobken, J.D.; Georgieff, M.K. Perinatal iron deficiency alters apical dendritic growth in hippocampal CA1 pyramidal neurons. Dev. Neurosci. 2003, 25, 412–420. [Google Scholar] [CrossRef] [PubMed]
- Jorgenson, L.A.; Sun, M.; O’Connor, M.; Georgieff, M.K. Fetal iron deficiency disrupts the maturation of synaptic function and efficacy in area CA1 of the developing rat hippocampus. Hippocampus 2005, 15, 1094–1102. [Google Scholar] [CrossRef]
- Rao, R.; Tkac, I.; Schmidt, A.T.; Georgieff, M.K. Fetal and neonatal iron deficiency causes volume loss and alters the neurochemical profile of the adult rat hippocampus. Nutr. Neurosci. 2011, 14, 59–65. [Google Scholar] [CrossRef]
- Clardy, S.L.; Wang, X.; Zhao, W.; Liu, W.; Chase, G.A.; Beard, J.L.; True Felt, B.; Connor, J.R. Acute and chronic effects of developmental iron deficiency on mRNA expression patterns in the brain. J. Neural Transm. Suppl. 2006, 173–196. [Google Scholar]
- Tran, P.V.; Kennedy, B.C.; Pisansky, M.T.; Won, K.; Gewirtz, J.C.; Simmons, R.A.; Georgieff, M.K. Prenatal choline supplementation diminishes early-life iron deficiency induced reprogramming of molecular networks associated with behavioral abnormalities in the adult rat hippocampus. J. Nutr. 2016, 146, 484–493. [Google Scholar] [CrossRef]
- Tran, P.V.; Carlson, E.S.; Fretham, S.J.; Georgieff, M.K. Early-life iron deficiency anemia alters neurotrophic factor expression and hippocampal neuron differentiation in male rats. J. Nutr. 2008, 138, 2495–2501. [Google Scholar] [CrossRef]
- Tran, P.V.; Kennedy, B.C.; Lien, Y.C.; Simmons, R.A.; Georgieff, M.K. Fetal iron deficiency induces chromatin remodeling at the Bdnf locus in adult rat hippocampus. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2015, 308, R276–R282. [Google Scholar] [CrossRef]
- Tahiliani, M.; Koh, K.P.; Shen, Y.; Pastor, W.A.; Bandukwala, H.; Brudno, Y.; Agarwal, S.; Iyer, L.M.; Liu, D.R.; Aravind, L.; et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 2009, 324, 930–935. [Google Scholar] [CrossRef]
- Hou, H.; Yu, H. Structural insights into histone lysine demethylation. Curr. Opin. Struct. Biol. 2010, 20, 739–748. [Google Scholar] [CrossRef] [Green Version]
- Moore, L.D.; Le, T.; Fan, G. DNA methylation and its basic function. Neuropsychopharmacology 2013, 38, 23–38. [Google Scholar] [CrossRef]
- Weber, M.; Hellmann, I.; Stadler, M.B.; Ramos, L.; Pääbo, S.; Rebhan, M.; Schübeler, D. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat. Genet. 2007, 39, 457–466. [Google Scholar] [CrossRef]
- Stevens, M.; Cheng, J.B.; Li, D.; Xie, M.; Hong, C.; Maire, C.L.; Ligon, K.L.; Hirst, M.; Marra, M.A.; Costello, J.F.; et al. Estimating absolute methylation levels at single-CpG resolution from methylation enrichment and restriction enzyme sequencing methods. Genome Res. 2013, 23, 1541–1553. [Google Scholar] [CrossRef] [Green Version]
- Condon, D.E.; Tran, P.V.; Lien, Y.C.; Schug, J.; Georgieff, M.K.; Simmons, R.A.; Won, K.J. Defiant: (DMRs: Easy, fast, identification and ANnoTation) identifies differentially Methylated regions from iron-deficient rat hippocampus. BMC Bioinf. 2018, 19, 31. [Google Scholar] [CrossRef]
- Fretham, S.J.; Carlson, E.S.; Georgieff, M.K. The role of iron in learning and memory. Adv. Nutr. 2011, 2, 112–121. [Google Scholar] [CrossRef]
- Tran, P.V.; Fretham, S.J.; Wobken, J.; Miller, B.S.; Georgieff, M.K. Gestational-neonatal iron deficiency suppresses and iron treatment reactivates IGF signaling in developing rat hippocampus. Am. J. Physiol. Endocrinol. Metab. 2012, 302, E316–E324. [Google Scholar] [CrossRef]
- Sheaffer, K.L.; Kim, R.; Aoki, R.; Elliott, E.N.; Schug, J.; Burger, L.; Schübeler, D.; Kaestner, K.H. DNA methylation is required for the control of stem cell differentiation in the small intestine. Genes Dev. 2014, 28, 652–664. [Google Scholar] [CrossRef] [Green Version]
- Carlson, E.S.; Stead, J.D.; Neal, C.R.; Petryk, A.; Georgieff, M.K. Perinatal iron deficiency results in altered developmental expression of genes mediating energy metabolism and neuronal morphogenesis in hippocampus. Hippocampus 2007, 17, 679–691. [Google Scholar] [CrossRef]
- Chilton, J.K. Molecular mechanisms of axon guidance. Dev. Biol. 2006, 292, 13–24. [Google Scholar] [CrossRef] [Green Version]
- Dickson, B.J. Molecular mechanisms of axon guidance. Science 2002, 298, 1959–1964. [Google Scholar] [CrossRef]
- Egea, J.; Klein, R. Bidirectional Eph-ephrin signaling during axon guidance. Trends Cell. Biol. 2007, 17, 230–238. [Google Scholar] [CrossRef]
- Rohani, N.; Canty, L.; Luu, O.; Fagotto, F.; Winklbauer, R. EphrinB/EphB signaling controls embryonic germ layer separation by contact-induced cell detachment. PLoS Biol. 2011, 9, e1000597. [Google Scholar] [CrossRef]
- Shamah, S.M.; Lin, M.Z.; Goldberg, J.L.; Estrach, S.; Sahin, M.; Hu, L.; Bazalakova, M.; Neve, R.L.; Corfas, G.; Debant, A.; et al. EphA receptors regulate growth cone dynamics through the novel guanine nucleotide exchange factor ephexin. Cell 2001, 105, 233–244. [Google Scholar] [CrossRef]
- Rex, C.S.; Chen, L.Y.; Sharma, A.; Liu, J.; Babayan, A.H.; Gall, C.M.; Lynch, G. Different Rho GTPase-dependent signaling pathways initiate sequential steps in the consolidation of long-term potentiation. J. Cell. Biol. 2009, 186, 85–97. [Google Scholar] [CrossRef]
- Malenka, R.C.; Nicoll, R.A. Long-term potentiation—A decade of progress? Science 1999, 285, 1870–1874. [Google Scholar] [CrossRef]
- Nicoll, R.A. A Brief History of Long-Term Potentiation. Neuron 2017, 93, 281–290. [Google Scholar] [CrossRef] [Green Version]
- Blattler, A.; Yao, L.; Witt, H.; Guo, Y.; Nicolet, C.M.; Berman, B.P.; Farnham, P.J. Global loss of DNA methylation uncovers intronic enhancers in genes showing expression changes. Genome Biol. 2014, 15, 469. [Google Scholar] [CrossRef]
- Singer, M.; Kosti, I.; Pachter, L.; Mandel-Gutfreund, Y. A diverse epigenetic landscape at human exons with implication for expression. Nucleic Acids Res. 2015, 43, 3498–3508. [Google Scholar] [CrossRef]
- Stephenson, D.T.; Coskran, T.M.; Kelly, M.P.; Kleiman, R.J.; Morton, D.; O’Neill, S.M.; Schmidt, C.J.; Weinberg, R.J.; Menniti, F.S. The distribution of phosphodiesterase 2A in the rat brain. Neuroscience 2012, 226, 145–155. [Google Scholar] [CrossRef] [Green Version]
- Fernández-Fernández, D.; Rosenbrock, H.; Kroker, K.S. Inhibition of PDE2A, but not PDE9A, modulates presynaptic short-term plasticity measured by paired-pulse facilitation in the CA1 region of the hippocampus. Synapse 2015, 69, 484–496. [Google Scholar] [CrossRef]
- Montague, P.; McCallion, A.S.; Davies, R.W.; Griffiths, I.R. Myelin-associated oligodendrocytic basic protein: A family of abundant CNS myelin proteins in search of a function. Dev. Neurosci. 2006, 28, 479–487. [Google Scholar] [CrossRef]
- Schäfer, I.; Müller, C.; Luhmann, H.J.; White, R. MOBP levels are regulated by Fyn kinase and affect the morphological differentiation of oligodendrocytes. J. Cell Sci. 2016, 129, 930–942. [Google Scholar] [CrossRef]
- Kuchler, K.; Daum, G.; Paltauf, F. Subcellular and submitochondrial localization of phospholipid-synthesizing enzymes in Saccharomyces cerevisiae. J. Bacteriol. 1986, 165, 901–910. [Google Scholar] [CrossRef] [Green Version]
- Nowicki, M.; Müller, F.; Frentzen, M. Cardiolipin synthase of Arabidopsis thaliana. FEBS Lett. 2005, 579, 2161–2165. [Google Scholar] [CrossRef]
- Qi, Y.; Kapterian, T.S.; Du, X.; Ma, Q.; Fei, W.; Zhang, Y.; Huang, X.; Dawes, I.W.; Yang, H. CDP-diacylglycerol synthases regulate the growth of lipid droplets and adipocyte development. J. Lipid Res. 2016, 57, 767–780. [Google Scholar] [CrossRef] [Green Version]
- Bastian, T.W.; von Hohenberg, W.C.; Georgieff, M.K.; Lanier, L.M. Chronic Energy Depletion due to Iron Deficiency Impairs Dendritic Mitochondrial Motility during Hippocampal Neuron Development. J. Neurosci. 2019, 39, 802–813. [Google Scholar] [CrossRef]
- Corapci, F.; Calatroni, A.; Kaciroti, N.; Jimenez, E.; Lozoff, B. Longitudinal evaluation of externalizing and internalizing behavior problems following iron deficiency in infancy. J. Pediatr. Psychol. 2010, 35, 296–305. [Google Scholar] [CrossRef]
- Lukowski, A.F.; Koss, M.; Burden, M.J.; Jonides, J.; Nelson, C.A.; Kaciroti, N.; Jimenez, E.; Lozoff, B. Iron deficiency in infancy and neurocognitive functioning at 19 years: Evidence of long-term deficits in executive function and recognition memory. Nutr. Neurosci. 2010, 13, 54–70. [Google Scholar] [CrossRef]
- Schmidt, R.J.; Tancredi, D.J.; Krakowiak, P.; Hansen, R.L.; Ozonoff, S. Maternal intake of supplemental iron and risk of autism spectrum disorder. Am. J. Epidemiol. 2014, 180, 890–900. [Google Scholar] [CrossRef]
- Connor, J.R.; Menzies, S.L. Relationship of iron to oligodendrocytes and myelination. Glia 1996, 17, 83–93. [Google Scholar] [CrossRef]
- Beard, J.L.; Wiesinger, J.A.; Connor, J.R. Pre- and postweaning iron deficiency alters myelination in Sprague-Dawley rats. Dev. Neurosci. 2003, 25, 308–315. [Google Scholar] [CrossRef]
- Tran, P.V.; Fretham, S.J.; Carlson, E.S.; Georgieff, M.K. Long-term reduction of hippocampal brain-derived neurotrophic factor activity after fetal-neonatal iron deficiency in adult rats. Pediatr. Res. 2009, 65, 493–498. [Google Scholar] [CrossRef]
- Thompson, R.F.; Fazzari, M.J.; Niu, H.; Barzilai, N.; Simmons, R.A.; Greally, J.M. Experimental intrauterine growth restriction induces alterations in DNA methylation and gene expression in pancreatic islets of rats. J. Biol. Chem. 2010, 285, 15111–15118. [Google Scholar] [CrossRef]
- Xiao, Y.; Camarillo, C.; Ping, Y.; Arana, T.B.; Zhao, H.; Thompson, P.M.; Xu, C.; Su, B.B.; Fan, H.; Ordonez, J.; et al. The DNA methylome and transcriptome of different brain regions in schizophrenia and bipolar disorder. PLoS ONE 2014, 9, e95875. [Google Scholar] [CrossRef]
- Li, M.; Gao, F.; Xia, Y.; Tang, Y.; Zhao, W.; Jin, C.; Luo, H.; Wang, J.; Li, Q.; Wang, Y. Filtrating colorectal cancer associated genes by integrated analyses of global DNA methylation and hydroxymethylation in cancer and normal tissue. Sci. Rep. 2016, 6, 31826. [Google Scholar] [CrossRef]
- Radford, E.J.; Ito, M.; Shi, H.; Corish, J.A.; Yamazawa, K.; Isganaitis, E.; Seisenberger, S.; Hore, T.A.; Reik, W.; Erkek, S.; et al. In utero effects. In utero undernourishment perturbs the adult sperm methylome and intergenerational metabolism. Science 2014, 345, 1255903. [Google Scholar] [CrossRef]
- Consortium, E.P. An integrated encyclopedia of DNA elements in the human genome. Nature 2012, 489, 57–74. [Google Scholar] [CrossRef]
- Pennacchio, L.A.; Ahituv, N.; Moses, A.M.; Prabhakar, S.; Nobrega, M.A.; Shoukry, M.; Minovitsky, S.; Dubchak, I.; Holt, A.; Lewis, K.D.; et al. In vivo enhancer analysis of human conserved non-coding sequences. Nature 2006, 444, 499–502. [Google Scholar] [CrossRef]
- Lev Maor, G.; Yearim, A.; Ast, G. The alternative role of DNA methylation in splicing regulation. Trends Genet. 2015, 31, 274–280. [Google Scholar] [CrossRef]
- Maunakea, A.K.; Nagarajan, R.P.; Bilenky, M.; Ballinger, T.J.; D’Souza, C.; Fouse, S.D.; Johnson, B.E.; Hong, C.; Nielsen, C.; Zhao, Y.; et al. Conserved role of intragenic DNA methylation in regulating alternative promoters. Nature 2010, 466, 253–257. [Google Scholar] [CrossRef]
- Oberdoerffer, S. A conserved role for intragenic DNA methylation in alternative pre-mRNA splicing. Transcription 2012, 3, 106–109. [Google Scholar] [CrossRef] [Green Version]
- Gelfman, S.; Cohen, N.; Yearim, A.; Ast, G. DNA-methylation effect on cotranscriptional splicing is dependent on GC architecture of the exon-intron structure. Genome Res. 2013, 23, 789–799. [Google Scholar] [CrossRef]
- Wan, J.; Oliver, V.F.; Zhu, H.; Zack, D.J.; Qian, J.; Merbs, S.L. Integrative analysis of tissue-specific methylation and alternative splicing identifies conserved transcription factor binding motifs. Nucleic Acids Res. 2013, 41, 8503–8514. [Google Scholar] [CrossRef] [Green Version]
- Ishihara, K.; Oshimura, M.; Nakao, M. CTCF-dependent chromatin insulator is linked to epigenetic remodeling. Mol. Cell. 2006, 23, 733–742. [Google Scholar] [CrossRef]
- Pikaart, M.J.; Recillas-Targa, F.; Felsenfeld, G. Loss of transcriptional activity of a transgene is accompanied by DNA methylation and histone deacetylation and is prevented by insulators. Genes Dev. 1998, 12, 2852–2862. [Google Scholar] [CrossRef] [Green Version]
- Ming, G.L.; Song, H.J.; Berninger, B.; Holt, C.E.; Tessier-Lavigne, M.; Poo, M.M. cAMP-dependent growth cone guidance by netrin-1. Neuron 1997, 19, 1225–1235. [Google Scholar] [CrossRef]
- Moore, S.W.; Kennedy, T.E. Protein kinase A regulates the sensitivity of spinal commissural axon turning to netrin-1 but does not switch between chemoattraction and chemorepulsion. J. Neurosci. 2006, 26, 2419–2423. [Google Scholar] [CrossRef]
- Sweatt, J.D. Toward a molecular explanation for long-term potentiation. Learn. Mem. 1999, 6, 399–416. [Google Scholar] [CrossRef]
- Thomas, M.J.; Moody, T.D.; Makhinson, M.; O’Dell, T.J. Activity-dependent beta-adrenergic modulation of low frequency stimulation induced LTP in the hippocampal CA1 region. Neuron 1996, 17, 475–482. [Google Scholar] [CrossRef]
- Bito, H.; Furuyashiki, T.; Ishihara, H.; Shibasaki, Y.; Ohashi, K.; Mizuno, K.; Maekawa, M.; Ishizaki, T.; Narumiya, S. A critical role for a Rho-associated kinase, p160ROCK, in determining axon outgrowth in mammalian CNS neurons. Neuron 2000, 26, 431–441. [Google Scholar] [CrossRef]
- Heasman, S.J.; Ridley, A.J. Mammalian Rho GTPases: New insights into their functions from in vivo studies. Nat. Rev. Mol. Cell. Biol. 2008, 9, 690–701. [Google Scholar] [CrossRef]
- Lakatosova, S.; Ostatnikova, D. Reelin and its complex involvement in brain development and function. Int. J. Biochem. Cell. Biol. 2012, 44, 1501–1504. [Google Scholar] [CrossRef]
- Weeber, E.J.; Beffert, U.; Jones, C.; Christian, J.M.; Forster, E.; Sweatt, J.D.; Herz, J. Reelin and ApoE receptors cooperate to enhance hippocampal synaptic plasticity and learning. J. Biol. Chem. 2002, 277, 39944–39952. [Google Scholar] [CrossRef]
- D’Arcangelo, G. Apoer2: A reelin receptor to remember. Neuron 2005, 47, 471–473. [Google Scholar] [CrossRef]
- Chen, Y.; Beffert, U.; Ertunc, M.; Tang, T.S.; Kavalali, E.T.; Bezprozvanny, I.; Herz, J. Reelin modulates NMDA receptor activity in cortical neurons. J. Neurosci. 2005, 25, 8209–8216. [Google Scholar] [CrossRef]
- Fatemi, S.H.; Kroll, J.L.; Stary, J.M. Altered levels of Reelin and its isoforms in schizophrenia and mood disorders. Neuroreport 2001, 12, 3209–3215. [Google Scholar] [CrossRef]
- Fatemi, S.H.; Stary, J.M.; Halt, A.R.; Realmuto, G.R. Dysregulation of Reelin and Bcl-2 proteins in autistic cerebellum. J. Autism Dev. Disord. 2001, 31, 529–535. [Google Scholar] [CrossRef]
- Li, E.; Zhang, Y. DNA methylation in mammals. Cold Spring Harb. Perspect. Biol. 2014, 6, a019133. [Google Scholar] [CrossRef]
- Zhao, B.; Yang, Y.; Wang, X.; Chong, Z.; Yin, R.; Song, S.H.; Zhao, C.; Li, C.; Huang, H.; Sun, B.F.; et al. Redox-active quinones induces genome-wide DNA methylation changes by an iron-mediated and Tet-dependent mechanism. Nucleic Acids Res. 2014, 42, 1593–1605. [Google Scholar] [CrossRef]
- Freudenberg, J.M.; Ghosh, S.; Lackford, B.L.; Yellaboina, S.; Zheng, X.; Li, R.; Cuddapah, S.; Wade, P.A.; Hu, G.; Jothi, R. Acute depletion of Tet1-dependent 5-hydroxymethylcytosine levels impairs LIF/Stat3 signaling and results in loss of embryonic stem cell identity. Nucleic Acids Res. 2012, 40, 3364–3377. [Google Scholar] [CrossRef]
- Zhu, X.; Girardo, D.; Govek, E.E.; John, K.; Mellén, M.; Tamayo, P.; Mesirov, J.P.; Hatten, M.E. Role of Tet1/3 Genes and Chromatin Remodeling Genes in Cerebellar Circuit Formation. Neuron 2016, 89, 100–112. [Google Scholar] [CrossRef]
- Branco, M.R.; Ficz, G.; Reik, W. Uncovering the role of 5-hydroxymethylcytosine in the epigenome. Nat. Rev. Genet. 2011, 13, 7–13. [Google Scholar] [CrossRef]
- Szulwach, K.E.; Li, X.; Li, Y.; Song, C.X.; Wu, H.; Dai, Q.; Irier, H.; Upadhyay, A.K.; Gearing, M.; Levey, A.I.; et al. 5-hmC-mediated epigenetic dynamics during postnatal neurodevelopment and aging. Nat. Neurosci. 2011, 14, 1607–1616. [Google Scholar] [CrossRef]
- Hahn, M.A.; Qiu, R.; Wu, X.; Li, A.X.; Zhang, H.; Wang, J.; Jui, J.; Jin, S.G.; Jiang, Y.; Pfeifer, G.P.; et al. Dynamics of 5-hydroxymethylcytosine and chromatin marks in Mammalian neurogenesis. Cell. Rep. 2013, 3, 291–300. [Google Scholar] [CrossRef]
- Al-Mahdawi, S.; Virmouni, S.A.; Pook, M.A. The emerging role of 5-hydroxymethylcytosine in neurodegenerative diseases. Front. Neurosci. 2014, 8, 397. [Google Scholar] [CrossRef] [Green Version]
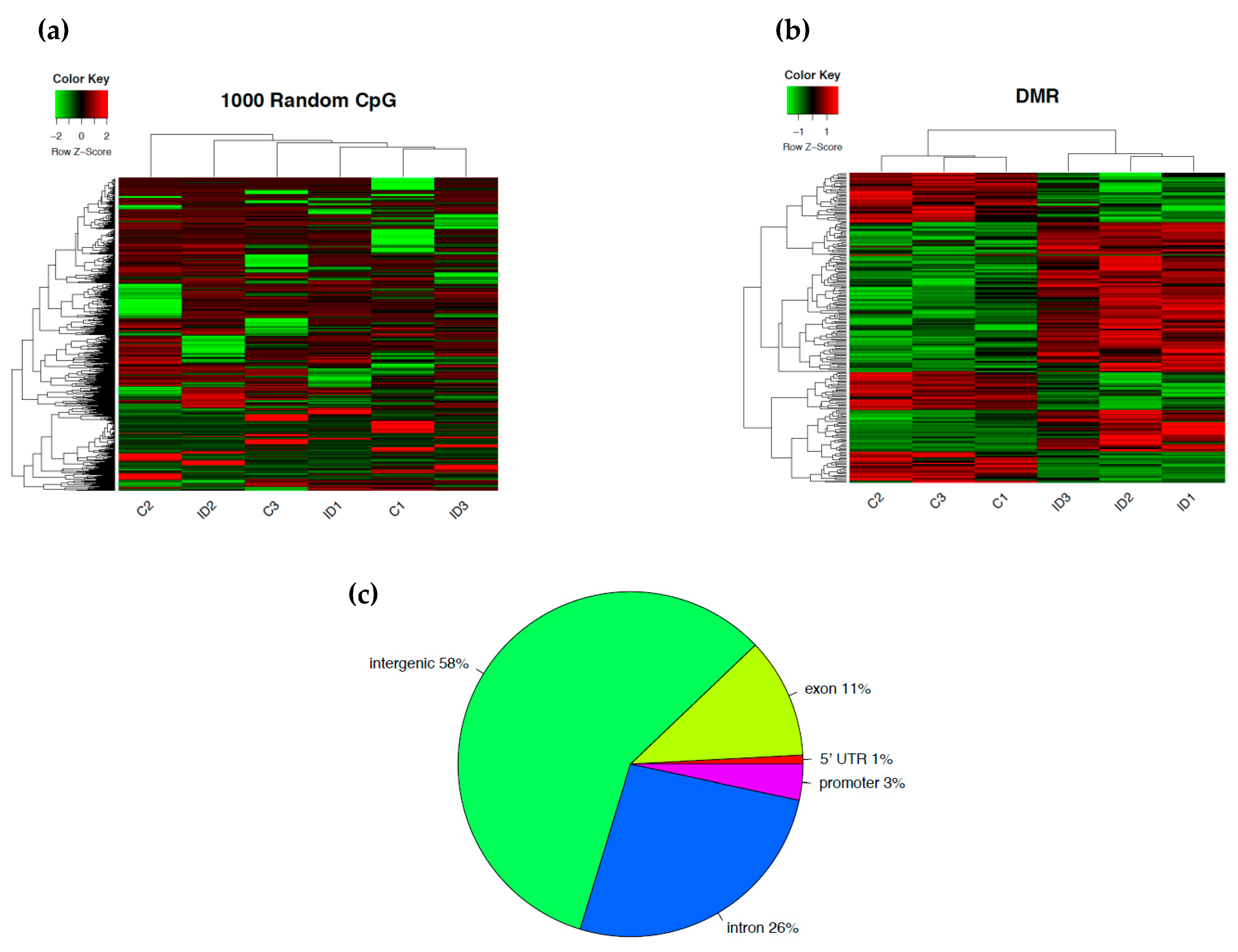
| Hypermethylation | Hypomethylation | ||||||
|---|---|---|---|---|---|---|---|
| Gene Name | #CpG | DMethylation(%) | q-value | Gene Name | #CpG | DMethylation(%) | q-value |
| Adamts19 | 5 | 58.5 | 0.016 | Abhd11 | 5 | −36.6 | 0.032 |
| Aebp1 | 5 | 10.5 | 0.118 | Adarb2 | 6 | −19.7 | 0.031 |
| Ak4 | 6 | 56.0 | 0.016 | Arhgap28 | 5 | −21.2 | 0.026 |
| Ankrd13a | 5 | 26.8 | 0.024 | Arhgef15 | 5 | −41.6 | 0.025 |
| Arf1 | 6 | 17.8 | 0.026 | Arhgef3 | 6 | −19.8 | 0.048 |
| Arhgap31 | 5 | 36.8 | 0.035 | Armc8 | 9 | −10.7 | 0.041 |
| Armc3 | 8 | 43.1 | 0.031 | Bag2 | 6 | −21.8 | 0.059 |
| B4galnt3 | 5 | 31.8 | 0.040 | Cds1 | 6 | −27.8 | 0.047 |
| Bcl11b | 6 | 44.7 | 0.035 | Commd1 | 6 | −40.4 | 0.016 |
| Btbd9 | 5 | 23.6 | 0.039 | Dip2a | 10 | −57.3 | 0.039 |
| Cacna1c | 5 | 27.9 | 0.032 | Dnaja2 | 6 | −39.0 | 0.037 |
| Camk2b | 9 | 19.3 | 0.032 | Dnpep | 5 | −14.3 | 0.051 |
| Capn12 | 7 | 21.5 | 0.024 | Dpf1 | 10 | −20.6 | 0.039 |
| Chd2 | 7 | 59.7 | 0.031 | Dpf3 | 6 | −13.3 | 0.018 |
| Clvs1 | 11 | 26.5 | 0.031 | Fkrp | 10 | −77.7 | 0.018 |
| Cog3 | 5 | 27.4 | 0.032 | Guca1a | 6 | −29.0 | 0.032 |
| Dgki | 5 | 51.3 | 0.026 | Hint1 | 8 | −25.1 | 0.025 |
| Ephb1 | 5 | 33.2 | 0.040 | Jak3 | 8 | −20.4 | 0.023 |
| Ezr | 5 | 51.9 | 0.018 | Kif26b | 6 | −44.2 | 0.031 |
| Fat3 | 9 | 50.1 | 0.018 | Klhl40 | 5 | −25.0 | 0.028 |
| Fig4 | 5 | 47.9 | 0.016 | Lims2 | 10 | −11.6 | 0.016 |
| Foxb2 | 5 | 32.6 | 0.039 | LOC691083 | 5 | −45.6 | 0.045 |
| Gucy2c | 9 | 86.6 | 0.016 | Mknk1 | 5 | −56.8 | 0.025 |
| Hip1r | 5 | 56.4 | 0.034 | Mobp | 5 | −48.9 | 0.048 |
| Iqcg | 9 | 52.6 | 0.020 | Ncf1 | 9 | −38.4 | 0.032 |
| Itsn1 | 5 | 29.8 | 0.032 | Pck1 | 5 | −48.2 | 0.023 |
| Jph3 | 8 | 25.1 | 0.024 | Pgm3 | 5 | −69.0 | 0.024 |
| Kank3 | 5 | 28.7 | 0.031 | Pon2 | 6 | −10.0 | 0.029 |
| Kctd15 | 6 | 55.1 | 0.032 | Ppp1r3b | 7 | −19.4 | 0.031 |
| Kctd6 | 7 | 27.3 | 0.016 | Rasd2 | 5 | −22.4 | 0.016 |
| Macrod1 | 5 | 35.4 | 0.033 | RGD735029 | 5 | −17.2 | 0.032 |
| Map3k11 | 10 | 12.9 | 0.016 | Sardh | 5 | −42.9 | 0.022 |
| Marveld2 | 6 | 17.9 | 0.018 | Sh3pxd2a | 5 | −39.4 | 0.016 |
| Mc3r | 5 | 42.2 | 0.016 | Slit3 | 5 | −41.5 | 0.029 |
| Mib1 | 6 | 48.9 | 0.032 | Smyd3 | 6 | −28.2 | 0.112 |
| Mogat1 | 5 | 27.4 | 0.035 | Ss18l1 | 6 | −18.6 | 0.042 |
| Mrpl19 | 6 | 23.9 | 0.059 | St8sia1 | 6 | −20.1 | 0.016 |
| Myo3b | 5 | 17.1 | 0.024 | Tal1 | 7 | −35.1 | 0.024 |
| Neto2 | 6 | 24.3 | 0.031 | Tmem120b | 5 | −37.7 | 0.024 |
| Olr987 | 5 | 21.1 | 0.016 | Tmem181 | 5 | −47.1 | 0.018 |
| Pabpn1l | 5 | 33.3 | 0.016 | Trrap | 5 | −28.2 | 0.023 |
| Pde2a | 5 | 28.6 | 0.025 | Usf2 | 10 | −18.3 | 0.016 |
| Pde6c | 6 | 46.1 | 0.024 | Ush1g | 9 | −17.2 | 0.037 |
| Ppp1r21 | 15 | 24.9 | 0.030 | Ust | 5 | −54.1 | 0.037 |
| Prkar1b | 5 | 40.2 | 0.024 | Wiz | 5 | −45.3 | 0.034 |
| Ptpn14 | 5 | 30.1 | 0.016 | ||||
| Rev3l | 5 | 16.7 | 0.023 | ||||
| Ric8b | 6 | 42.0 | 0.025 | ||||
| Riok2 | 5 | 48.0 | 0.031 | ||||
| Sbk1 | 6 | 39.5 | 0.026 | ||||
| Scrt2 | 6 | 36.8 | 0.038 | ||||
| Slc38a1 | 5 | 38.2 | 0.029 | ||||
| Slc5a1 | 7 | 37.4 | 0.026 | ||||
| Snurf | 5 | 34.7 | 0.001 | ||||
| Spon1 | 8 | 73.6 | 0.031 | ||||
| Srgap2 | 5 | 28.6 | 0.026 | ||||
| Tbc1d20 | 6 | 13.5 | 0.042 | ||||
| Tenm2 | 5 | 42.5 | 0.016 | ||||
| Tfap2b | 5 | 22.6 | 0.035 | ||||
| Tgif2 | 5 | 32.2 | 0.017 | ||||
| Tnni1 | 31 | 29.8 | 0.016 | ||||
| Unc93b1 | 5 | 54.8 | 0.020 | ||||
| Usp36 | 21 | 28.8 | 0.043 | ||||
| Ingenuity Canonical Pathways | p-Value | Differentially Methylated Genes in the Pathway |
|---|---|---|
| Nitric Oxide Signaling in the Cardiovascular System | 0.002 | CACNA1C, PRKAR1B, PDE2A, GUCY2C |
| Cardiac β-Adrenergic Signaling | 0.005 | CACNA1C, PRKAR1B, PDE2A, PDE6C |
| cAMP-Mediated Signaling | 0.005 | CAMK2B, MC3R, PRKAR1B, PDE2A, PDE6C |
| Axonal Guidance Signaling | 0.006 | ARHGEF15, ITSN1, SLIT3, MKNK1, PRKAR1B, EPHB1, SRGAP2 |
| Relaxin Signaling | 0.007 | PRKAR1B, PDE2A, GUCY2C, PDE6C |
| Reelin Signaling in Neurons | 0.010 | ARHGEF15, ARHGEF3, MAP3K11 |
| G-Protein Coupled Receptor Signaling | 0.011 | CAMK2B, MC3R, PRKAR1B, PDE2A, PDE6C |
| Protein Kinase A Signaling | 0.013 | CAMK2B, PTPN14, TNNI1, PRKAR1B, PDE2A, PDE6C |
| Synaptic Long-Term Potentiation | 0.021 | CAMK2B, CACNA1C, PRKAR1B |
| Signaling by Rho Family GTPases | 0.034 | ARHGEF15, ARHGEF3, MAP3K11, EZR |
| Methylome Analysis | Microarray Analysis | |||
|---|---|---|---|---|
| Ingenuity Canonical Pathways | p-value | Differentially Methylated Genes | p-value | Differentially Expressed Genes |
| Nitric Oxide Signaling in the Cardiovascular System | 0.002 | CACNA1C, PRKAR1B, PDE2A, GUCY2C | 0.000 | ITPR2, PIK3R3, KDR, PTPN11, PRKAA1, GUCY2D, ITPR1, CAMK4, PRKAG1, PDE2A, PDGFC |
| Cellular Effects of Sildenafil (Viagra) | 0.004 | CACNA1C, PRKAR1B, PDE2A, GUCY2C | 0.000 | MYH3, CACNG8, ITPR2, ADCY3, GPR37, GUCY2D, ITPR1, ADCY2, PLCE1, CAMK4, PRKAG1, PDE2A |
| Cardiac β-Adrenergic Signaling | 0.005 | CACNA1C, PRKAR1B, PDE2A, PDE6C | 0.036 | ADCY3, PKIG, ADCY2, PRKAG1, PDE2A, PPP2R2A, PPP1R11 |
| cAMP-Mediated Signaling | 0.005 | CAMK2B, MC3R, PRKAR1B, PDE2A, PDE6C | 0.000 | GABBR1, CHRM3, CAMK4, VIPR1, PDE2A, Htr5b, CHRM2, CNGA2, CAMK2A, GNAI3, ADCY3, HRH3, PKIG, ADCY2, LHCGR, OPRM1, GRM6 |
| Axonal Guidance Signaling | 0.006 | ARHGEF15, ITSN1, SLIT3, MKNK1, PRKAR1B, EPHB1, SRGAP2 | 0.003 | CXCL12, PIK3R3, TUBB, EPHA3, ROBO1, PLCE1, DPYSL5, RTN4R, RTN4, GNAI3, FZD4, PDGFC, BAIAP2, SEMA4F, CXCR4, NRAS, CFL1, PTPN11, NTRK2, PRKAG1 |
| Relaxin Signaling | 0.007 | PRKAR1B, PDE2A, GUCY2C, PDE6C | 0.008 | PIK3R3, ADCY3, PTPN11, GUCY2D, ADCY2, PRKAG1, PDE2A, NFKBIA, GNAI3 |
| Reelin Signaling in Neurons | 0.010 | ARHGEF15, ARHGEF3, MAP3K11 | 0.004 | PAFAH1B1, PIK3R3, PTPN11, APP, MAPT, ARHGEF9, APBB1 |
| G-Protein Coupled Receptor Signaling | 0.011 | CAMK2B, MC3R, PRKAR1B, PDE2A, PDE6C | 0.000 | PIK3R3, GABBR1, CHRM3, CAMK4, VIPR1, PDE2A, Htr5b, NFKBIA, CHRM2, CAMK2A, GNAI3, NRAS, PDPK1, ADCY3, HRH3, PTPN11, ADCY2, PRKAG1, GRM5, LHCGR, OPRM1, GRM6 |
| Protein Kinase A Signaling | 0.013 | CAMK2B, Ptpn14, TNNI1, PRKAR1B, PDE2A, PDE6C | 0.000 | ITPR2, PLCE1, NFKBIA, CNGA2, GNAI3, PYGB, ADCY3, PTPN11, ITPR1, ADCY2, PTPRF, TGFBR1, PPP1R1B, YWHAB, PPP1R11, DUSP12, PTPRN, CDC25A, PTPN2, PTPRO, H3F3A/H3F3B, CAMK4, PDE2A, PTPN23, CAMK2A, BAD, DUSP5, PTPN12, PRKAG1 |
| Breast Cancer Regulation by Stathmin1 | 0.017 | CAMK2B, ARHGEF15, ARHGEF3, PRKAR1B | 0.000 | ITPR2, PIK3R3, TUBB, CAMK4, PPP2R2A, CAMK2A, GNAI3, STMN1, NRAS, ADCY3, PTPN11, ITPR1, ADCY2, ARHGEF9, PRKAG1, PPP1R11 |
| Synaptic Long-Term Potentiation | 0.021 | CAMK2B, CACNA1C, PRKAR1B | 0.000 | NRAS, ITPR2, GRINA, ITPR1, PLCE1, CAMK4, PRKAG1, GRM5, GRIN1, CAMK2A, GRM6, PPP1R11 |
| Gustation Pathway | 0.023 | PRKAR1B, PDE2A, PDE6C | 0.000 | CACNG8, ITPR2, ADCY3, CACNB4, CACNA2D1, P2RX5, ITPR1, ADCY2, PRKAG1, PDE2A, P2RY1, CACNA1H |
| Sperm Motility | 0.023 | MAP3K11, PRKAR1B, PDE2A | 0.002 | ITPR2, PAFAH1B1, ITPR1, PLCE1, CAMK4, PRKAG1, PDE2A, CNGA2, CACNA1H |
| GNRH Signaling | 0.032 | CAMK2B, MAP3K11, PRKAR1B | 0.000 | CACNG8, ITPR2, CACNB4, CAMK4, CAMK2A, GNAI3, CACNA1H, NRAS, ADCY3, CACNA2D1, GNRHR, ITPR1, ADCY2, PRKAG1 |
| Signaling by Rho Family GTPases | 0.034 | ARHGEF15, ARHGEF3, MAP3K11, EZR | 0.010 | BAIAP2, CFL1, RHOT2, PIK3R3, PTPN11, RHOB, CDH1, ARHGEF9, PLD1, RHOV, GNAI3, STMN1 |
| Molecular Mechanisms of Cancer | 0.042 | CAMK2B, ARHGEF15, ARHGEF3, JAK3, PRKAR1B | 0.000 | RASGRF1, RHOT2, PIK3R3, CDC25A, CASP9, NFKBIA, CAMK2A, BAD, GNAI3, FZD4, NCSTN, NRAS, RALBP1, ADCY3, PTPN11, RHOB, HIF1A, ADCY2, CASP3, TGFBR1, CDH1, ARHGEF9, PRKAG1, RHOV |
| Melatonin Signaling | 0.048 | CAMK2B, PRKAR1B | 0.021 | PLCE1, CAMK4, PRKAG1, CAMK2A, GNAI3 |
| Ephrin B Signaling | 0.049 | ITSN1, EPHB1 | 0.022 | CXCL12, CXCR4, CFL1, CAP1, GNAI3 |
| Category | Diseases or Functions Annotation | p-value | Differentially Methylated Genes | p-value | Number of Genes |
|---|---|---|---|---|---|
| Cell-To-Cell Signaling | Synaptic Depression/Neurotransmission | 1.65E-04 | CAMK2B, ARF1, ITSN1, DGKI, PRKAR1B, EPHB1 | 1.94E-10 | 21 |
| Nervous System Development and Function | Neuritogenesis/Extension of Neurites | 8.40E-03 | CAMK2B, ST8SIA1, ITSN1, SS18L1, BCL11B, EZR, SLIT3, UST, EPHB1, SRGAP2 | 4.92E-16 | 62 |
| Behavior | Locomotion | 3.09E-04 | RASD2, HINT1, MC3R, BTBD9, NCF1, CACNA1C, JPH3, FIG4, TAL1 | 1.08E-13 | 40 |
| Learning | 2.22E-02 | CAMK2B, NCF1, BTBD9, DGKI, CACNA1C, PRKAR1B, JPH3 | 3.51E-21 | 57 | |
| Neurological Disease | Cell Death of Cerebral Cortex Cells | 1.33E-02 | ST8SIA1, ITSN1, MAP3K11, NCF1, SH3PXD2A | 8.55E-14 | 32 |
| Movement Disorder | 4.68E-02 | CAMK2B, AEBP1, CDS1, ST8SIA1, HINT1, BCL11B, TFAP2B, PDE6C, USP36, RASD2, MC3R, BTBD9, CACNA1C | 5.58E-32 | 117 | |
| Lipid Metabolism | Quantity of Sphingolipid/Steroid | 2.73E-03 | ST8SIA1, HINT1, BCL11B, PON2 | 4.24E-09 | 40 |
| Molecular Transport | Quantity of Heavy Metal | 1.13E-02 | ARF1, USF2, COMMD1 | 4.32E-19 | 58 |
| Transport of Molecule | 1.92E-02 | SLC5A1, SLC38A1 | 5.41E-31 | 144 |
| Gene Name | Symbol | ∆ Methylation (%) | CpGs Location | FC (ID/IS) | Location | Type(s) |
|---|---|---|---|---|---|---|
| Phosphodiesterase 2A | Pde2a | 28.6 | Intron 2 | 1.16 | Plasma Membrane | enzyme |
| Myelin-associated oligodendrocyte basic protein | Mobp | −48.9 | Intron 2 | 1.37 | Cytoplasm | other |
| CDP-diacylglycerol synthase 1 | Cds1 | −27.8 | Intron 11 | 1.23 | Endoplasmic reticulum & mitochondria | enzyme |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lien, Y.-C.; Condon, D.E.; Georgieff, M.K.; Simmons, R.A.; Tran, P.V. Dysregulation of Neuronal Genes by Fetal-Neonatal Iron Deficiency Anemia Is Associated with Altered DNA Methylation in the Rat Hippocampus. Nutrients 2019, 11, 1191. https://0-doi-org.brum.beds.ac.uk/10.3390/nu11051191
Lien Y-C, Condon DE, Georgieff MK, Simmons RA, Tran PV. Dysregulation of Neuronal Genes by Fetal-Neonatal Iron Deficiency Anemia Is Associated with Altered DNA Methylation in the Rat Hippocampus. Nutrients. 2019; 11(5):1191. https://0-doi-org.brum.beds.ac.uk/10.3390/nu11051191
Chicago/Turabian StyleLien, Yu-Chin, David E Condon, Michael K Georgieff, Rebecca A Simmons, and Phu V Tran. 2019. "Dysregulation of Neuronal Genes by Fetal-Neonatal Iron Deficiency Anemia Is Associated with Altered DNA Methylation in the Rat Hippocampus" Nutrients 11, no. 5: 1191. https://0-doi-org.brum.beds.ac.uk/10.3390/nu11051191




