The Preparation of Ginsenoside Rg5, Its Antitumor Activity against Breast Cancer Cells and Its Targeting of PI3K
Abstract
:1. Introduction
2. Materials and Methods
2.1. Experimental Materials
2.2. Preparation of Ginsenoside Rg5
2.3. Cell Culture
2.4. Cell Viability Assay
2.5. Clone Formation Assay
2.6. Acridine Orange/Ethidium Bromide (AO/EB) Staining Assay
2.7. Flow Cytometric Analysis of Apoptosis
2.8. Measurement of Mitochondrial Membrane Potential (MMP)
2.9. Western Blot Assay
2.10. Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
2.11. MitoTracker Green Assay
2.12. Transmission Electronic Microscopy (TEM)
2.13. Immunofluorescence Microscopy
2.14. Molecular Docking
2.15. ADMET Prediction
2.16. Statistical Analysis
3. Results
3.1. Comparison of the Cytotoxicities of Rb1, R-Rg3, S-Rg3, and Rg5 in Various Tumor Cells
3.2. Rg5 Inhibits Breast Cancer Cell Viability
3.3. Rg5 Induces Caspase-Dependent Apoptosis in Breast Cancer Cells
3.4. Rg5 Induces Apoptosis via the Mitochondria-Mediated Pathway
3.5. Rg5 Triggers Autophagy, Which Promotes Cell Death in Breast Cancer Cells
3.6. Rg5 Augments the Fusion of Autophagosomes and Lysosomes in Breast Cancer Cells
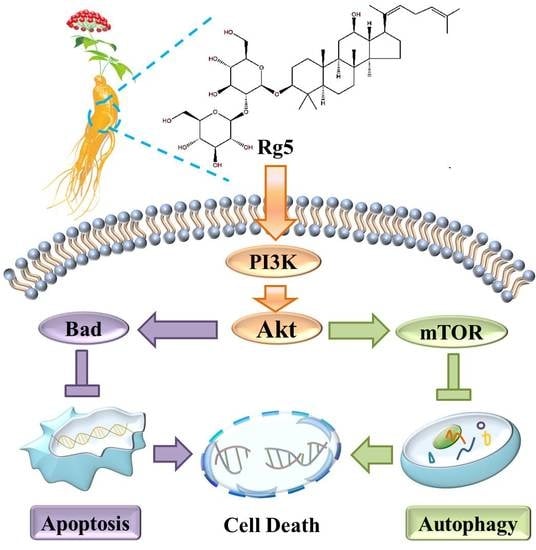
3.7. Rg5 Triggers Apoptosis and Autophagy by Blocking the PI3K Pathway in Breast Cancer Cells
3.8. Molecular Docking Study and ADMET Prediction
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Africander, D.; Storbeck, K.H. Steroid metabolism in breast cancer: Where are we and what are we missing? Mol. Cell. Endocrinol. 2017, 466, 86–97. [Google Scholar] [CrossRef]
- Siegel, R.L.; KD, M.; Jemal, A. Cancer statistics, 2018. Ca A Cancer J. Clin. 2018, 68, 7. [Google Scholar] [CrossRef]
- Yewale, C.; Baradia, D.; Patil, S.; Bhatt, P.; Amrutiya, J.; Gandhi, R.; Kore, G.; Misra, A. Docetaxel loaded immunonanoparticles delivery in EGFR overexpressed breast carcinoma cells. J. Drug Deliv. Sci. Technol. 2018, 45, 334–345. [Google Scholar] [CrossRef]
- Oberoi, H.S.; Nukolova, N.V.; Kabanov, A.V.; Bronich, T.K. Nanocarriers for delivery of platinum anticancer drugs. Adv. Drug Deliv. Rev. 2013, 65, 1667–1685. [Google Scholar] [CrossRef] [Green Version]
- Gong, C.; Hu, C.; Gu, F.; Xia, Q.; Yao, C.; Zhang, L.; Qiang, L.; Gao, S.; Gao, Y. Co-delivery of autophagy inhibitor ATG7 siRNA and docetaxel for breast cancer treatment. J Control Release 2017, 266, 272–286. [Google Scholar] [CrossRef]
- Markman, M. Safety issues in using complementary and alternative medicine. J. Clin. Oncol. 2002, 20, 39S–41S. [Google Scholar]
- Li, L.; Lee, S.J.; Yuan, Q.P.; Wan, T.I.; Sun, C.K.; Han, N.S. Production of bioactive ginsenoside Rg3(S) and compound K using recombinant Lactococcus lactis. J. Ginseng Res. 2017, 42, 412–418. [Google Scholar] [CrossRef]
- Park, C.S.; Yoo, M.H.; Noh, K.H.; Oh, D.K. Biotransformation of ginsenosides by hydrolyzing the sugar moieties of ginsenosides using microbial glycosidases. Appl. Microbiol. Biotechnol. 2010, 87, 9–19. [Google Scholar] [CrossRef]
- Li, W.; Gu, C.; Zhang, H.; Awang, D.V.; Fitzloff, J.F.; Fong, H.H.; Van Breemen, R.B. Use of high-performance liquid chromatography-tandem mass spectrometry to distinguish Panax ginseng C. A. Meyer (Asian ginseng) and Panax quinquefolius L. (North American ginseng). Anal. Chem. 2000, 72, 5417–5422. [Google Scholar] [CrossRef]
- Qi, L.W.; Wang, C.Z.; Yuan, C.S. Ginsenosides from American ginseng: Chemical and pharmacological diversity. Phytochemistry 2011, 72, 689–699. [Google Scholar] [CrossRef] [Green Version]
- Lee, H.; Lee, S.; Jeong, D.; Sun, J.K. Ginsenoside Rh2 epigenetically regulates cell-mediated immune pathway to inhibit proliferation of MCF-7 breast cancer cells. J. Ginseng Res. 2017, 42, 455–462. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.J.; Ji, S.L.; Lee, E.; Lim, T.G.; Byun, S. The ginsenoside metabolite compound K inhibits hormone-independent breast cancer through downregulation of cyclin D1. J. Funct. Foods 2018, 46, 159–166. [Google Scholar] [CrossRef]
- Eun-Hwa, P.; Young-Joo, K.; Noriko, Y.; Soon-Hye, P.; Ho-Kyong, K.; Hyuk-Jai, J.; Hoon, K.J.; Gab Jin, C.; Jungyeob, H.; Ki Sung, K. Stereospecific anticancer effects of ginsenoside Rg3 epimers isolated from heat-processed American ginseng on human gastric cancer cell. J. Ginseng Res. 2014, 38, 22–27. [Google Scholar]
- Jiang, J.; Yuan, Z.; Sun, Y.; Bu, Y.; Li, W.; Fei, Z. Ginsenoside Rg3 enhances the anti-proliferative activity of erlotinib in pancreatic cancer cell lines by downregulation of EGFR/PI3K/Akt signaling pathway. Biomed. Pharmacother. 2017, 96, 619–625. [Google Scholar] [CrossRef] [PubMed]
- Duan, Z.; Deng, J.; Dong, Y.; Zhu, C.; Li, W.; Fan, D. Anticancer effects of ginsenoside Rk3 on non-small cell lung cancer cells: In vitro and in vivo. Food Funct. 2017, 8, 3723–3736. [Google Scholar] [CrossRef]
- Wu, Q.; Deng, J.; Fan, D.; Duan, Z.; Zhu, C.; Fu, R.; Wang, S. Ginsenoside Rh4 induces apoptosis and autophagic cell death through activation of the ROS/JNK/p53 pathway in colorectal cancer cells. Biochem. Pharmacol. 2017, 148, 64–74. [Google Scholar] [CrossRef]
- Liang, L.; Tao, H.E.; Tingwei, D.U.; Fan, Y.; Chen, D.; Wang, Y. Ginsenoside-Rg5 induces apoptosis and DNA damage in human cervical cancer cells. Mol. Med. Rep. 2015, 11, 940–946. [Google Scholar] [CrossRef] [Green Version]
- Chang, K.H.; Mi, N.J.; Kim, K.T.; Paik, H.D. Evaluation of glucosidases of Aspergillus niger strain comparing with other glucosidases in transformation of ginsenoside Rb1 to ginsenosides Rg3. J. Ginseng Res. 2014, 38, 47–51. [Google Scholar] [CrossRef] [Green Version]
- Nagalingam, A.; Tighiouart, M.; Ryden, L.; Joseph, L.; Landberg, G.; Saxena, N.K.; Sharma, D. Med1 plays a critical role in the development of tamoxifen resistance. Carcinogenesis 2012, 33, 918–930. [Google Scholar] [CrossRef] [Green Version]
- Chung, S.J.; Nagaraju, G.P.; Nagalingam, A.; Muniraj, N.; Sharma, D. ADIPOQ/adiponectin induces cytotoxic autophagy in breast cancer cells through STK11/LKB1-mediated activation of the AMPK-ULK1 axis. Autophagy 2017, 13, 1–18. [Google Scholar] [CrossRef]
- Kimura, S.; Noda, T.; Yoshimori, T. Dissection of the Autophagosome Maturation Process by a Novel Reporter Protein, Tandem Fluorescent-Tagged LC3. Autophagy 2007, 3, 452–460. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pei, J.; Xie, J.; Ran, Y.; Zhao, L.; Gang, D.; Wang, Z.; Wei, X. Enzymatic transformation of ginsenoside Rb1 to ginsenoside 20(S)-Rg3 by GH3 β-glucosidase from Thermotoga thermarum DSM 5069 T. J. Mol. Catal. B-Enzym. 2015, 113, 104–109. [Google Scholar] [CrossRef]
- Yan-Cong, L.; Shu-Ming, H.; Zhi-Xu, H.; Minghua, L.; Yinxue, Y.; Jian-Xin, P.; Xueji, Z.; Kevin, C.; Qingyu, Z.; Wei, D. Plumbagin induces apoptotic and autophagic cell death through inhibition of the PI3K/Akt/mTOR pathway in human non-small cell lung cancer cells. Cancer Lett. 2014, 344, 239–259. [Google Scholar]
- Roos, W.P.; Thomas, A.D.; Kaina, B. DNA damage and the balance between survival and death in cancer biology. Nat. Rev. Cancer 2015, 16, 20. [Google Scholar] [CrossRef]
- Bai, L.; Wang, S. Targeting Apoptosis Pathways for New Cancer Therapeutics. Annu. Rev. Med. 2013, 65, 139–155. [Google Scholar] [CrossRef]
- Qin, N.; Lu, S.; Chen, N.; Chen, C.; Xie, Q.; Wei, X.; Ye, F.; He, J.; Li, Y.; Chen, L. Yulangsan polysaccharide inhibits 4T1 breast cancer cell proliferation and induces apoptosis in vitro and in vivo. Int. J. Biol. Macromol. 2019, 121, 971–980. [Google Scholar] [CrossRef]
- Fu, S.; Chen, X.; Lo, H.W.; Lin, J. Combined bazedoxifene and paclitaxel treatments inhibit cell viability, cell migration, colony formation, and tumor growth and induce apoptosis in breast cancer. Cancer Lett. 2019, 448, 11–19. [Google Scholar] [CrossRef]
- Gibson, C.J.; Davids, M.S. BCL-2 Antagonism to Target the Intrinsic Mitochondrial Pathway of Apoptosis. Clin. Cancer Res. 2015, 21, 5021–5029. [Google Scholar] [CrossRef] [Green Version]
- Lu, Y.; Shihua, W.; Qiuhui, Z.; Feiyan, L.; Ping, W. 23,24-Dihydrocucurbitacin B induces G2/M cell-cycle arrest and mitochondria-dependent apoptosis in human breast cancer cells (Bcap37). Cancer Lett. 2007, 256, 267–278. [Google Scholar]
- Patel, O.P.; Arun, A.; Singh, P.K.; Saini, D.; Karade, S.S.; Chourasia, M.K.; Konwar, R.; Yadav, P.P. Pyranocarbazole derivatives as potent anti-cancer agents triggering tubulin polymerization stabilization induced activation of caspase-dependent apoptosis and downregulation of Akt/mTOR in breast cancer cells. Eur. J. Med. Chem. 2019, 167, 226–244. [Google Scholar] [CrossRef]
- Patricia, B.; Fulvio, R.; Patrice, C. Emerging regulation and functions of autophagy. Nat. Cell Biol. 2013, 15, 713–720. [Google Scholar]
- Lamb, C.A.; Tamotsu, Y.; Tooze, S.A. The autophagosome: Origins unknown, biogenesis complex. Nat. Rev. Mol. Cell Biol. 2013, 14, 759–774. [Google Scholar] [CrossRef] [PubMed]
- Yuchen, F.; Ding, H.; Zhiyuan, Y.; Klionsky, D.J. The machinery of macroautophagy. Cell Res. 2014, 24, 24. [Google Scholar]
- Sahani, M.H.; Itakura, E.; Mizushima, N. Expression of the autophagy substrate SQSTM1/p62 is restored during prolonged starvation depending on transcriptional upregulation and autophagy-derived amino acids. Autophagy 2014, 10, 431–441. [Google Scholar] [CrossRef] [Green Version]
- Siddharth, S.; Muniraj, N.; Saxena, N.; Sharma, D. Concomitant Inhibition of Cytoprotective Autophagy Augments the Efficacy of Withaferin A in Hepatocellular Carcinoma. Cancers 2019, 11, 453. [Google Scholar] [CrossRef] [Green Version]
- Muniraj, N.; Siddharth, S.; Nagalingam, A.; Walker, A.; Woo, J.; Győrffy, B.; Gabrielson, E.; Saxena, N.K.; Sharma, D. Withaferin A inhibits lysosomal activity to block autophagic flux and induces apoptosis via energetic impairment in breast cancer cells. Carcinogenesis 2019, 40, 1110–1120. [Google Scholar] [CrossRef]
- Mizushima, N.; Yoshimori, T.; Levine, B. Methods in Mammalian Autophagy Research. Cell 2010, 140, 1–326. [Google Scholar] [CrossRef] [Green Version]
- Eisenberglerner, A.; Bialik, S.; Simon, H.U.; Kimchi, A. Life and death partners: Apoptosis, autophagy and the cross-talk between them. Cell Death Differ. 2009, 16, 966. [Google Scholar] [CrossRef]
- Wang, X.; Wei, S.; Zhao, Y.; Shi, C.; Liu, P.; Zhang, C.; Lei, Y.; Zhang, B.; Bai, B.; Huang, Y. Anti-proliferation of breast cancer cells with itraconazole: Hedgehog pathway inhibition induces apoptosis and autophagic cell death. Cancer Lett. 2016, 385, 128–136. [Google Scholar] [CrossRef]
- Zhang, J.; Li, L.; Peng, Y.; Chen, Y.; Lv, X.; Li, S.; Qin, X.; Yang, H.; Wu, C.; Liu, Y. Surface chemistry induces mitochondria-mediated apoptosis of breast cancer cells via PTEN/PI3K/AKT signaling pathway. Biochimica Biophysica Acta 2017, 1865, 172–185. [Google Scholar] [CrossRef]
- Yang, S.X.; Polley, E.; Lipkowitz, S. New insights on PI3K/AKT pathway alterations and clinical outcomes in breast cancer. Cancer Treat Rev. 2016, 45, 87–96. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jia, L.; Huang, S.; Yin, X.; Zan, Y.; Guo, Y.; Han, L. Quercetin suppresses the mobility of breast cancer by suppressing glycolysis through Akt-mTOR pathway mediated autophagy induction. Life Sci. 2018, 208, 123–130. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Li, R.; Shi, W.; Tao, J.; Wang, Y.; Cong, L.; Qu, X. Silencing of MicroRNA-21 confers the sensitivity to tamoxifen and fulvestrant by enhancing autophagic cell death through inhibition of the PI3K-AKT-mTOR pathway in breast cancer cells. Biomed. Pharmacother. 2016, 77, 37–44. [Google Scholar] [CrossRef] [PubMed]
- Zhou, B.H.; Tan, P.P.; Jia, L.S.; Zhao, W.P.; Wang, J.C.; Wang, H.W. PI3K/AKT signaling pathway involvement in fluoride-induced apoptosis in C2C12 cells. Chemosphere 2018, 199, 297–302. [Google Scholar] [CrossRef] [PubMed]
- Yanfei, M.; Huadong, Q.; Yunfu, C. MiR-34a targets GAS1 to promote cell proliferation and inhibit apoptosis in papillary thyroid carcinoma via PI3K/Akt/Bad pathway. Biochem. Biophys. Res. Commun. 2013, 441, 958–963. [Google Scholar]









| Antibody | Working Dilution | Molecular Weight (KDa) | Cat. Number |
|---|---|---|---|
| Cleaved caspase-3 | 1:1000 | 17 | AB29034 |
| Cleaved caspase-8 | 1:1000 | 47 | AB40502 |
| Cleaved caspase-9 | 1:1000 | 17 | AB40503 |
| Cleaved PARP | 1:1000 | 85 | AB29033 |
| Bax | 1:5000 | 21 | 50599-2-Ig |
| Bcl-2 | 1:2000 | 26 | 12789-1-AP |
| Atg5 | 1:1000 | 55 | 10181-2-AP |
| Atg12 | 1:1000 | 48 | 11122-1-AP |
| Beclin-1 | 1:2000 | 60 | 11306-1-AP |
| p62 | 1:2000 | 62 | 66184-1-Ig |
| LC3B | 1:4000 | 16, 18 | AB51520 |
| Cytochrome c | 1:1000 | 14 | 4280 |
| β-actin | 1:4000 | 42 | 20536-1-AP |
| PI3K | 1:1000 | 110 | 20584-1-AP |
| phospho-PI3K | 1:1000 | 85 | ABP50495 |
| AKT | 1:1000 | 60 | AB21054 |
| phospho-AKT | 1:1000 | 60 | AB11054 |
| mTOR | 1:1000 | 289 | AB21214 |
| phospho-mTOR | 1:1000 | 289 | AB11221 |
| Bad | 1:2000 | 18 | 10435-1-AP |
| phospho-Bad | 1:1000 | 23 | AB11068 |
| Goat anti-Rabbit IgG | 1:20000 | — | A21020 |
| Genes | Forward Primer (5′–3′) | Reverse Primer (5′–3′) |
|---|---|---|
| β-Actin | ttccagccttgcttcctg | tacttgcgcttgggagga |
| Bax | cccccgagaggtcttttt | tcccggaggaagtccaat |
| Bcl-2 | aatgtgcccagcctcttg | tctgttgcccaactgcaa |
| LC3B | atgttgccacctcccaaa | tccagcacgagttcacga |
| P62 | agcgggcatcagtttgag | gccctcctttccgatgat |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Y.; Fan, D. The Preparation of Ginsenoside Rg5, Its Antitumor Activity against Breast Cancer Cells and Its Targeting of PI3K. Nutrients 2020, 12, 246. https://0-doi-org.brum.beds.ac.uk/10.3390/nu12010246
Liu Y, Fan D. The Preparation of Ginsenoside Rg5, Its Antitumor Activity against Breast Cancer Cells and Its Targeting of PI3K. Nutrients. 2020; 12(1):246. https://0-doi-org.brum.beds.ac.uk/10.3390/nu12010246
Chicago/Turabian StyleLiu, Yannan, and Daidi Fan. 2020. "The Preparation of Ginsenoside Rg5, Its Antitumor Activity against Breast Cancer Cells and Its Targeting of PI3K" Nutrients 12, no. 1: 246. https://0-doi-org.brum.beds.ac.uk/10.3390/nu12010246