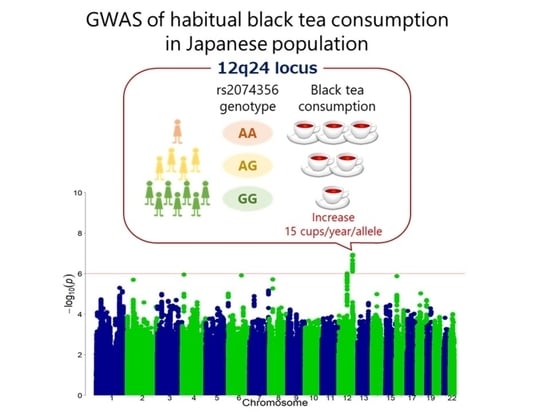
A Genome-Wide Association Study Identifies the Association between the 12q24 Locus and Black Tea Consumption in Japanese Populations
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Design
2.2. Black Tea Consumption
2.3. Adjustment Variables
2.4. DNA Sampling, Genotype, Quality Control, and Genotype Imputation
2.5. Genome-Wide Association and Meta-Analysis
2.6. Confounding Factor Adjustment and Subgroup Analysis
3. Results
3.1. Research Flow and Characteristics of the Study Participants
3.2. Discovery GWAS
3.3. Replication Stage and Meta-Analysis
3.4. Adjustment for Potential Confounding Factors
3.5. Subgroup Analysis According to Sex and Age
4. Discussion
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Leung, L.K.; Su, Y.; Chen, R.; Zhang, Z.; Huang, Y.; Chen, Z.-Y. Theaflavins in Black Tea and Catechins in Green Tea Are Equally Effective Antioxidants. J. Nutr. 2001, 131, 2248–2251. [Google Scholar] [CrossRef]
- Cheang, W.S.; Ngai, C.Y.; Tam, Y.Y.; Tian, X.Y.; Wong, W.T.; Zhang, Y.; Lau, C.W.; Chen, Z.Y.; Bian, Z.X.; Huang, Y.; et al. Black tea protects against hypertension-associated endothelial dysfunction through alleviation of endoplasmic reticulum stress. Sci. Rep. 2015, 5, 10340. [Google Scholar] [CrossRef]
- Cameron, A.R.; Anton, S.; Melville, L.; Houston, N.P.; Dayal, S.; McDougall, G.J.; Stewart, D.; Rena, G. Black tea polyphenols mimic insulin/insulin-like growth factor-1 signalling to the longevity factor FOXO1a. Aging Cell 2008, 7, 69–77. [Google Scholar] [CrossRef] [PubMed]
- Gothandam, K.; Ganesan, V.S.; Ayyasamy, T.; Ramalingam, S. Antioxidant potential of theaflavin ameliorates the activities of key enzymes of glucose metabolism in high fat diet and streptozotocin-induced diabetic rats. Redox Rep. Commun. Free Radic. Res. 2019, 24, 41–50. [Google Scholar] [CrossRef] [Green Version]
- Arab, L.; Liu, W.; Elashoff, D. Green and black tea consumption and risk of stroke: A meta-analysis. Stroke 2009, 40, 1786–1792. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Robino, A.; Concas, M.P.; Catamo, E.; Gasparini, P. A Brief Review of Genetic Approaches to the Study of Food Preferences: Current Knowledge and Future Directions. Nutrients 2019, 11, 1735. [Google Scholar] [CrossRef] [Green Version]
- Matoba, N.; Akiyama, M.; Ishigaki, K.; Kanai, M.; Takahashi, A.; Momozawa, Y.; Ikegawa, S.; Ikeda, M.; Iwata, N.; Hirata, M.; et al. GWAS of 165,084 Japanese individuals identified nine loci associated with dietary habits. Nat. Hum. Behav. 2020, 4, 308–316. [Google Scholar] [CrossRef] [PubMed]
- Igarashi, M.; Nogawa, S.; Kawafune, K.; Hachiya, T.; Takahashi, S.; Jia, H.; Saito, K.; Kato, H. Identification of the 12q24 locus associated with fish intake frequency by genome-wide meta-analysis in Japanese populations. Genes Nutr. 2019, 14, 21. [Google Scholar] [CrossRef] [PubMed]
- Jorgenson, E.; Thai, K.K.; Hoffmann, T.J.; Sakoda, L.C.; Kvale, M.N.; Banda, Y.; Schaefer, C.; Risch, N.; Mertens, J.; Weisner, C.; et al. Genetic contributors to variation in alcohol consumption vary by race/ethnicity in a large multi-ethnic genome-wide association study. Mol. Psychiatry 2017, 22, 1359–1367. [Google Scholar] [CrossRef] [Green Version]
- Quillen, E.E.; Chen, X.-D.; Almasy, L.; Yang, F.; He, H.; Li, X.; Wang, X.-Y.; Liu, T.-Q.; Hao, W.; Deng, H.-W.; et al. ALDH2 is associated to alcohol dependence and is the major genetic determinant of “daily maximum drinks” in a GWAS study of an isolated rural Chinese sample. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2014, 165B, 103–110. [Google Scholar] [CrossRef] [Green Version]
- Yang, X.; Lu, X.; Wang, L.; Chen, S.; Li, J.; Cao, J.; Chen, J.; Hao, Y.; Li, Y.; Zhao, L.; et al. Common variants at 12q24 are associated with drinking behavior in Han Chinese. Am. J. Clin. Nutr. 2013, 97, 545–551. [Google Scholar] [CrossRef] [PubMed]
- Jia, H.; Nogawa, S.; Kawafune, K.; Hachiya, T.; Takahashi, S.; Igarashi, M.; Saito, K.; Kato, H. GWAS of habitual coffee consumption reveals a sex difference in the genetic effect of the 12q24 locus in the Japanese population. BMC Genet. 2019, 20, 61. [Google Scholar] [CrossRef] [PubMed]
- Nakagawa-Senda, H.; Hachiya, T.; Shimizu, A.; Hosono, S.; Oze, I.; Watanabe, M.; Matsuo, K.; Ito, H.; Hara, M.; Nishida, Y.; et al. A genome-wide association study in the Japanese population identifies the 12q24 locus for habitual coffee consumption: The J-MICC Study. Sci. Rep. 2018, 8, 1493. [Google Scholar] [CrossRef]
- Pirastu, N.; Kooyman, M.; Robino, A.; van der Spek, A.; Navarini, L.; Amin, N.; Karssen, L.C.; Van Duijn, C.M.; Gasparini, P. Non-additive genome-wide association scan reveals a new gene associated with habitual coffee consumption. Sci. Rep. 2016, 6, 31590. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kawafune, K.; Hachiya, T.; Nogawa, S.; Takahashi, S.; Jia, H.; Saito, K.; Kato, H. Strong association between the 12q24 locus and sweet taste preference in the Japanese population revealed by genome-wide meta-analysis. J. Hum. Genet. 2020. [Google Scholar] [CrossRef] [PubMed]
- Price, A.L.; Patterson, N.J.; Plenge, R.M.; Weinblatt, M.E.; Shadick, N.A.; Reich, D. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 2006, 38, 904–909. [Google Scholar] [CrossRef]
- Yamaguchi-Kabata, Y.; Nakazono, K.; Takahashi, A.; Saito, S.; Hosono, N.; Kubo, M.; Nakamura, Y.; Kamatani, N. Japanese population structure, based on SNP genotypes from 7003 individuals compared to other ethnic groups: Effects on population-based association studies. Am. J. Hum. Genet. 2008, 83, 445–456. [Google Scholar] [CrossRef] [Green Version]
- Hachiya, T.; Komaki, S.; Hasegawa, Y.; Ohmomo, H.; Tanno, K.; Hozawa, A.; Tamiya, G.; Yamamoto, M.; Ogasawara, K.; Nakamura, M.; et al. Genome-wide meta-analysis in Japanese populations identifies novel variants at the TMC6–TMC8 and SIX3–SIX2 loci associated with HbA1c. Sci. Rep. 2017, 7, 16147. [Google Scholar] [CrossRef]
- Shimanoe, C.; Hachiya, T.; Hara, M.; Nishida, Y.; Tanaka, K.; Sutoh, Y.; Shimizu, A.; Hishida, A.; Kawai, S.; Okada, R.; et al. A genome-wide association study of coping behaviors suggests FBXO45 is associated with emotional expression. Genes Brain Behav. 2019, 18, e12481. [Google Scholar] [CrossRef]
- Chang, C.C.; Chow, C.C.; Tellier, L.C.; Vattikuti, S.; Purcell, S.M.; Lee, J.J. Second-generation PLINK: Rising to the challenge of larger and richer datasets. GigaScience 2015, 4, 7. [Google Scholar] [CrossRef]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.R.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.W.; Daly, M.J.; et al. PLINK: A Tool Set for Whole-Genome Association and Population-Based Linkage Analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- The Genomes Project Consortium; Auton, A.; Abecasis, G.R.; Altshuler, D.M.; Durbin, R.M.; Abecasis, G.R.; Bentley, D.R.; Chakravarti, A.; Clark, A.G.; Donnelly, P.; et al. A global reference for human genetic variation. Nature 2015, 526, 68. [Google Scholar] [CrossRef] [Green Version]
- Loh, P.-R.; Danecek, P.; Palamara, P.F.; Fuchsberger, C.; Reshef, Y.A.; Finucane, H.K.; Schoenherr, S.; Forer, L.; McCarthy, S.; Abecasis, G.R.; et al. Reference-based phasing using the Haplotype Reference Consortium panel. Nat. Genet. 2016, 48, 1443–1448. [Google Scholar] [CrossRef] [Green Version]
- Das, S.; Forer, L.; Schönherr, S.; Sidore, C.; Locke, A.E.; Kwong, A.; Vrieze, S.I.; Chew, E.Y.; Levy, S.; McGue, M.; et al. Next-generation genotype imputation service and methods. Nat. Genet. 2016, 48, 1284–1287. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Willer, C.J.; Li, Y.; Abecasis, G.R. METAL: Fast and efficient meta-analysis of genomewide association scans. Bioinformatics (Oxford) 2010, 26, 2190–2191. [Google Scholar] [CrossRef]
- Kim, Y.J.; Go, M.J.; Hu, C.; Hong, C.B.; Kim, Y.K.; Lee, J.Y.; Hwang, J.-Y.; Oh, J.H.; Kim, D.-J.; Kim, N.H.; et al. Large-scale genome-wide association studies in East Asians identify new genetic loci influencing metabolic traits. Nat. Genet. 2011, 43, 990–995. [Google Scholar] [CrossRef]
- Zhang, H.; Mo, X.B.; Xu, T.; Bu, X.Q.; Lei, S.F.; Zhang, Y.H. Novel Genes Affecting Blood Pressure Detected Via Gene-Based Association Analysis. G3 (Bethesda) 2015, 5, 1035–1042. [Google Scholar] [CrossRef] [Green Version]
- Wen, W.; Zheng, W.; Okada, Y.; Takeuchi, F.; Tabara, Y.; Hwang, J.Y.; Dorajoo, R.; Li, H.; Tsai, F.J.; Yang, X.; et al. Meta-analysis of genome-wide association studies in East Asian-ancestry populations identifies four new loci for body mass index. Hum. Mol. Genet. 2014, 23, 5492–5504. [Google Scholar] [CrossRef] [Green Version]
- Cornelis, M.C.; Monda, K.L.; Yu, K.; Paynter, N.; Azzato, E.M.; Bennett, S.N.; Berndt, S.I.; Boerwinkle, E.; Chanock, S.; Chatterjee, N.; et al. Genome-wide meta-analysis identifies regions on 7p21 (AHR) and 15q24 (CYP1A2) as determinants of habitual caffeine consumption. PLoS Genet. 2011, 7, e1002033. [Google Scholar] [CrossRef]
- Josse, A.R.; Da Costa, L.A.; Campos, H.; El-Sohemy, A. Associations between polymorphisms in the AHR and CYP1A1-CYP1A2 gene regions and habitual caffeine consumption. Am. J. Clin. Nutr. 2012, 96, 665–671. [Google Scholar] [CrossRef] [Green Version]
- Higdon, J.V.; Frei, B. Coffee and Health: A Review of Recent Human Research. Crit. Rev. Food Sci. Nutr. 2006, 46, 101–123. [Google Scholar] [CrossRef]
- Jin, T.; Youn, J.; Kim, A.N.; Kang, M.; Kim, K.; Sung, J.; Lee, J.E. Interactions of Habitual Coffee Consumption by Genetic Polymorphisms with the Risk of Prediabetes and Type 2 Diabetes Combined. Nutrients 2020, 12, 2228. [Google Scholar] [CrossRef]
- Fang, C.M.; Roy, S.; Nielsen, E.; Paul, M.; Maul, R.; Paun, A.; Koentgen, F.; Raval, F.M.; Szomolanyi-Tsuda, E.; Pitha, P.M. Unique contribution of IRF-5-Ikaros axis to the B-cell IgG2a response. Genes Immun. 2012, 13, 421–430. [Google Scholar] [CrossRef] [Green Version]
- Drabsch, T.; Holzapfel, C. A Scientific Perspective of Personalised Gene-Based Dietary Recommendations for Weight Management. Nutrients 2019, 11, 617. [Google Scholar] [CrossRef] [Green Version]





| Parameters | Discovery | Replication |
|---|---|---|
| N | 12,140 | 118 |
| Female (%) | 46.8 | 45.8 |
| Age, years (mean ± SD) | 50.3 ± 13.2 | 49.0 ± 12.3 |
| Black tea consumption, cups/day (mean ± SD) | 0.20 ± 0.51 | 0.15 ± 0.41 |
| Drinking frequency, times/week (mean ± SD) | 2.21 ± 2.67 | 2.63 ± 2.76 |
| Alcohol consumption, g/day (means ± SD) | 7.10 ± 11.94 | 9.87 ± 15.02 |
| Coffee consumption, cups/day (mean ± SD) | 1.70 ± 1.50 | 1.64 ± 1.28 |
| Sweet taste preference, (mean ± SD) | 3.74 ± 0.90 | 3.72 ± 0.89 |
| BMI, kg/m2 (mean ± SD) | 23.1 ± 3.7 | 23.9 ± 4.0 |
| SNP | Chr | Position | Gene | EA | NEA | Population | EAF | Beta | SE (Beta) | pAssociation |
|---|---|---|---|---|---|---|---|---|---|---|
| rs2074356 | 12 | 112645401 | HECTD4 | T | C | Discovery | 0.244 | 0.040 | 0.008 | 1.2 × 10−7 |
| Replication | 0.174 | 0.184 | 0.070 | 0.01 | ||||||
| Meta-analysis | 0.243 | 0.042 | 0.008 | 2.4 × 10−8 | ||||||
| rs144504271 | 12 | 112627350 | HECTD4 | A | G | Discovery | 0.264 | 0.041 | 0.008 | 1.2 × 10−7 |
| Replication | 0.190 | 0.183 | 0.072 | 0.01 | ||||||
| Meta-analysis | 0.263 | 0.042 | 0.008 | 3.1 × 10−8 | ||||||
| rs12231737 | 12 | 112574616 | TRAFD1 | T | C | Discovery | 0.266 | 0.041 | 0.008 | 1.4 × 10−7 |
| Replication | 0.192 | 0.187 | 0.072 | 0.01 | ||||||
| Meta-analysis | 0.265 | 0.043 | 0.008 | 4.3 × 10−8 | ||||||
| rs116873087 | 12 | 112511913 | NAA25 | C | G | Discovery | 0.263 | 0.041 | 0.008 | 2.2 × 10−7 |
| Replication | 0.189 | 0.186 | 0.074 | 0.01 | ||||||
| Meta-analysis | 0.262 | 0.043 | 0.008 | 5.8 × 10−8 | ||||||
| rs11066132 | 12 | 112468206 | NAA25 | T | C | Discovery | 0.261 | 0.040 | 0.008 | 3.2 × 10−7 |
| Replication | 0.188 | 0.188 | 0.074 | 0.01 | ||||||
| Meta-analysis | 0.260 | 0.042 | 0.008 | 9.6 × 10−8 | ||||||
| rs78069066 | 12 | 112337924 | MAPKAPK5 TMEM116 | A | G | Discovery | 0.267 | 0.039 | 0.008 | 3.5 × 10−7 |
| Replication | 0.189 | 0.177 | 0.072 | 0.01 | ||||||
| Meta-analysis | 0.266 | 0.040 | 0.008 | 1.1 × 10−7 | ||||||
| rs4646776 | 12 | 112230019 | ALDH2 | C | G | Discovery | 0.264 | 0.037 | 0.007 | 4.4 × 10−7 |
| Replication | 0.187 | 0.171 | 0.070 | 0.02 | ||||||
| Meta-analysis | 0.263 | 0.039 | 0.007 | 1.4 × 10−7 | ||||||
| rs671 | 12 | 112241766 | ALDH2 | A | G | Discovery | 0.264 | 0.037 | 0.007 | 4.5 × 10−7 |
| Replication | 0.186 | 0.171 | 0.070 | 0.02 | ||||||
| Meta-analysis | 0.263 | 0.039 | 0.007 | 1.5 × 10−7 | ||||||
| rs11066001 | 12 | 112119171 | BRAP | C | T | Discovery | 0.262 | 0.038 | 0.008 | 6.2 × 10−7 |
| Replication | 0.185 | 0.175 | 0.072 | 0.02 | ||||||
| Meta-analysis | 0.261 | 0.039 | 0.008 | 1.6 × 10−7 | ||||||
| rs11066015 | 12 | 112168009 | ACAD10 | A | G | Discovery | 0.263 | 0.037 | 0.007 | 5.7 × 10−7 |
| Replication | 0.187 | 0.171 | 0.070 | 0.02 | ||||||
| Meta-analysis | 0.262 | 0.038 | 0.007 | 1.8 × 10−7 | ||||||
| rs3782886 | 12 | 112110489 | BRAP | C | T | Discovery | 0.264 | 0.037 | 0.008 | 7.9 × 10−7 |
| Replication | 0.186 | 0.175 | 0.072 | 0.02 | ||||||
| Meta-analysis | 0.263 | 0.039 | 0.008 | 2.6 × 10−7 | ||||||
| rs1981764 | 12 | 68217659 | DYRK2– IFNG | G | A | Discovery | 0.123 | 0.049 | 0.010 | 9.9 × 10−7 |
| Replication | 0.119 | −0.021 | 0.087 | 0.81 | ||||||
| Meta-analysis | 0.127 | 0.048 | 0.010 | 1.4 × 10−6 |
| Adjustment Variables | Beta | SE (Beta) | p Association |
|---|---|---|---|
| Age, Sex, Population structure (5 PCs) | 0.040 | 0.008 | 1.2 × 10−7 |
| Age, Sex, Population structure (5 PCs), Drinking frequency | 0.033 | 0.008 | 3.8 × 10−5 |
| Age, Sex, Population structure (5 PCs), Alcohol consumption | 0.037 | 0.008 | 3.1 × 10−6 |
| Age, Sex, Population structure (5 PCs), Coffee consumption | 0.045 | 0.008 | 9.9 × 10−9 |
| Age, Sex, Population structure (5 PCs), Sweet preference | 0.039 | 0.008 | 3.0 × 10−7 |
| Age, Sex, Population structure (5 PCs), BMI | 0.040 | 0.008 | 9.9 × 10−8 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Furukawa, K.; Igarashi, M.; Jia, H.; Nogawa, S.; Kawafune, K.; Hachiya, T.; Takahashi, S.; Saito, K.; Kato, H. A Genome-Wide Association Study Identifies the Association between the 12q24 Locus and Black Tea Consumption in Japanese Populations. Nutrients 2020, 12, 3182. https://0-doi-org.brum.beds.ac.uk/10.3390/nu12103182
Furukawa K, Igarashi M, Jia H, Nogawa S, Kawafune K, Hachiya T, Takahashi S, Saito K, Kato H. A Genome-Wide Association Study Identifies the Association between the 12q24 Locus and Black Tea Consumption in Japanese Populations. Nutrients. 2020; 12(10):3182. https://0-doi-org.brum.beds.ac.uk/10.3390/nu12103182
Chicago/Turabian StyleFurukawa, Kyohei, Maki Igarashi, Huijuan Jia, Shun Nogawa, Kaoru Kawafune, Tsuyoshi Hachiya, Shoko Takahashi, Kenji Saito, and Hisanori Kato. 2020. "A Genome-Wide Association Study Identifies the Association between the 12q24 Locus and Black Tea Consumption in Japanese Populations" Nutrients 12, no. 10: 3182. https://0-doi-org.brum.beds.ac.uk/10.3390/nu12103182