Metabolomic and Metataxonomic Fingerprinting of Human Milk Suggests Compositional Stability over a Natural Term of Breastfeeding to 24 Months
Abstract
:1. Introduction
2. Materials and Methods
2.1. Participant Recruitment
2.2. Sample Collection and Handling
2.3. Sample Processing and Total Volume and Fat Determination
2.4. Total DNA Extraction from Human Milk Samples and Metataxonomic Analysis
2.5. Metabolic Fingerprinting Using Laser Assisted Rapid Evaporative Ionisation Mass Spectrometry (LA-REIMS)
2.6. Correlation Analysis of Multi-Omic Datasets
2.7. Statistical Analysis
2.8. Availability of Raw Data
3. Results and Discussion
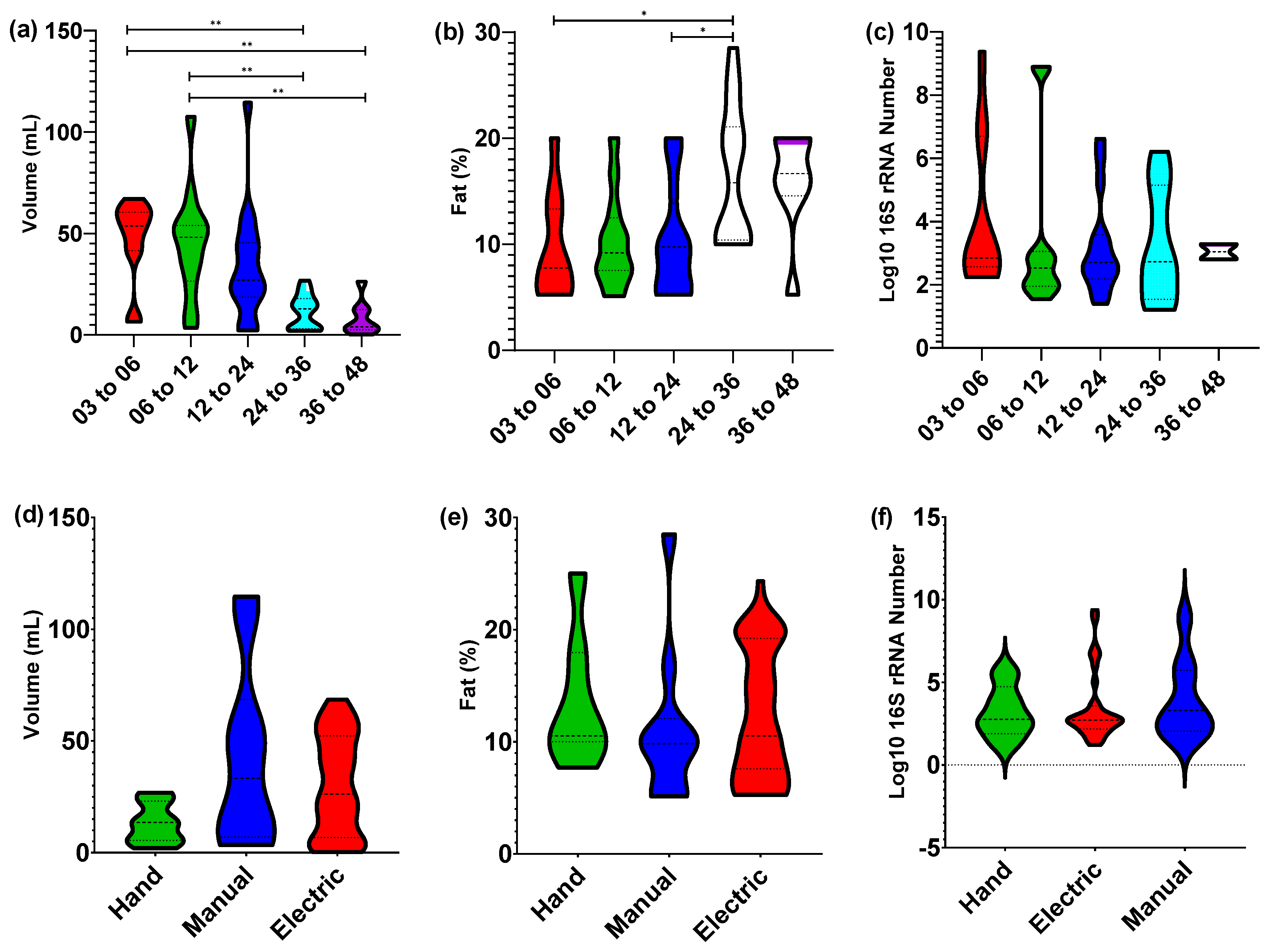
3.1. Nursling Age Correlates Only with Expression Volume
3.2. Core bacterial Microbiome Exists across Lactation Period
3.3. Detection of Metabolite Features in Human Milk Using LA-REIMS
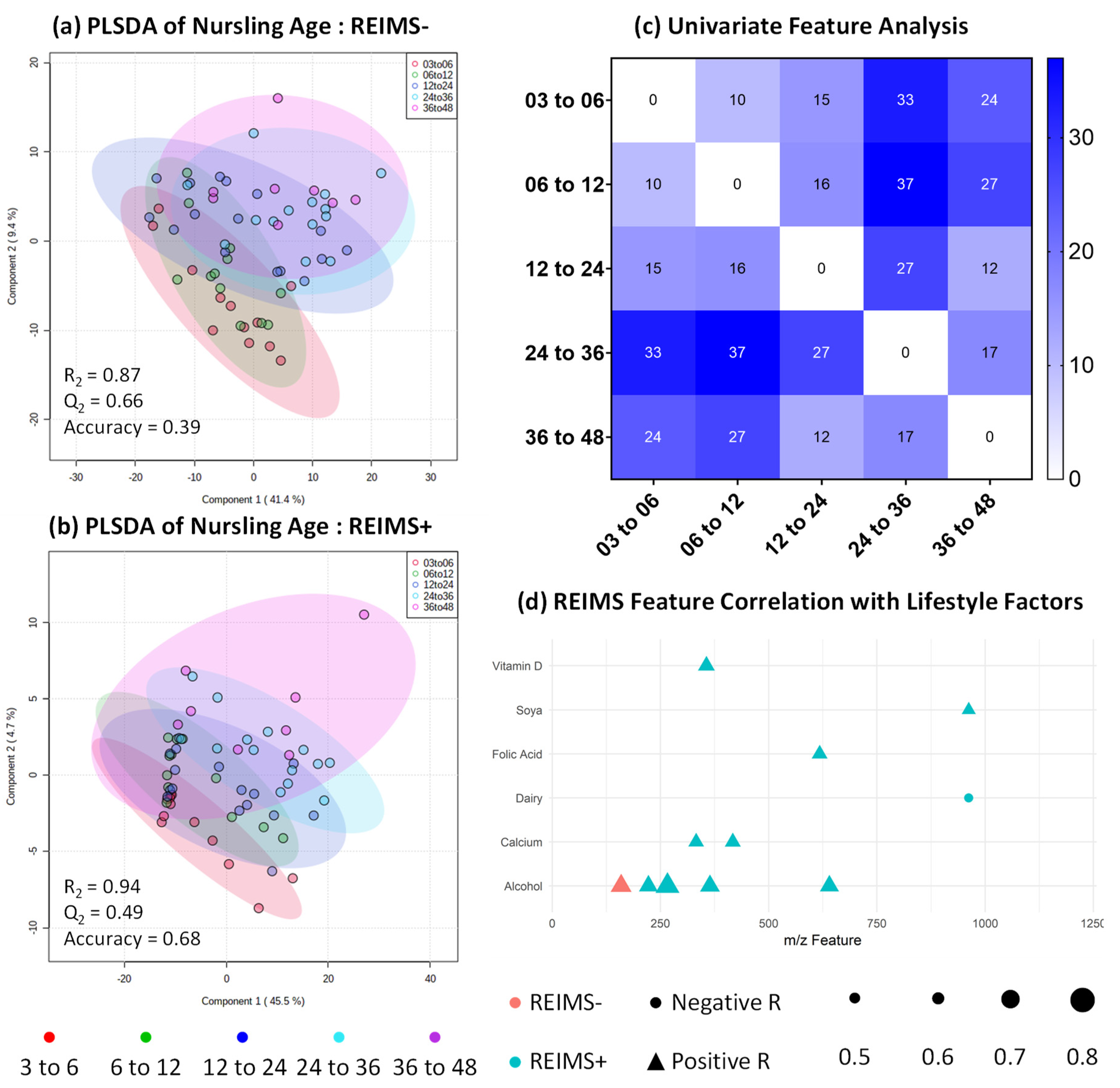
3.4. Metabolomic Fingerprinting Indicates Subtle Shifts beyond 2 Years
3.5. Potential Confounding Variables on Human Milk Composition
3.6. Human Milk Shows Compositional Stability against Self-reported Lifestyle and Dietary Factors
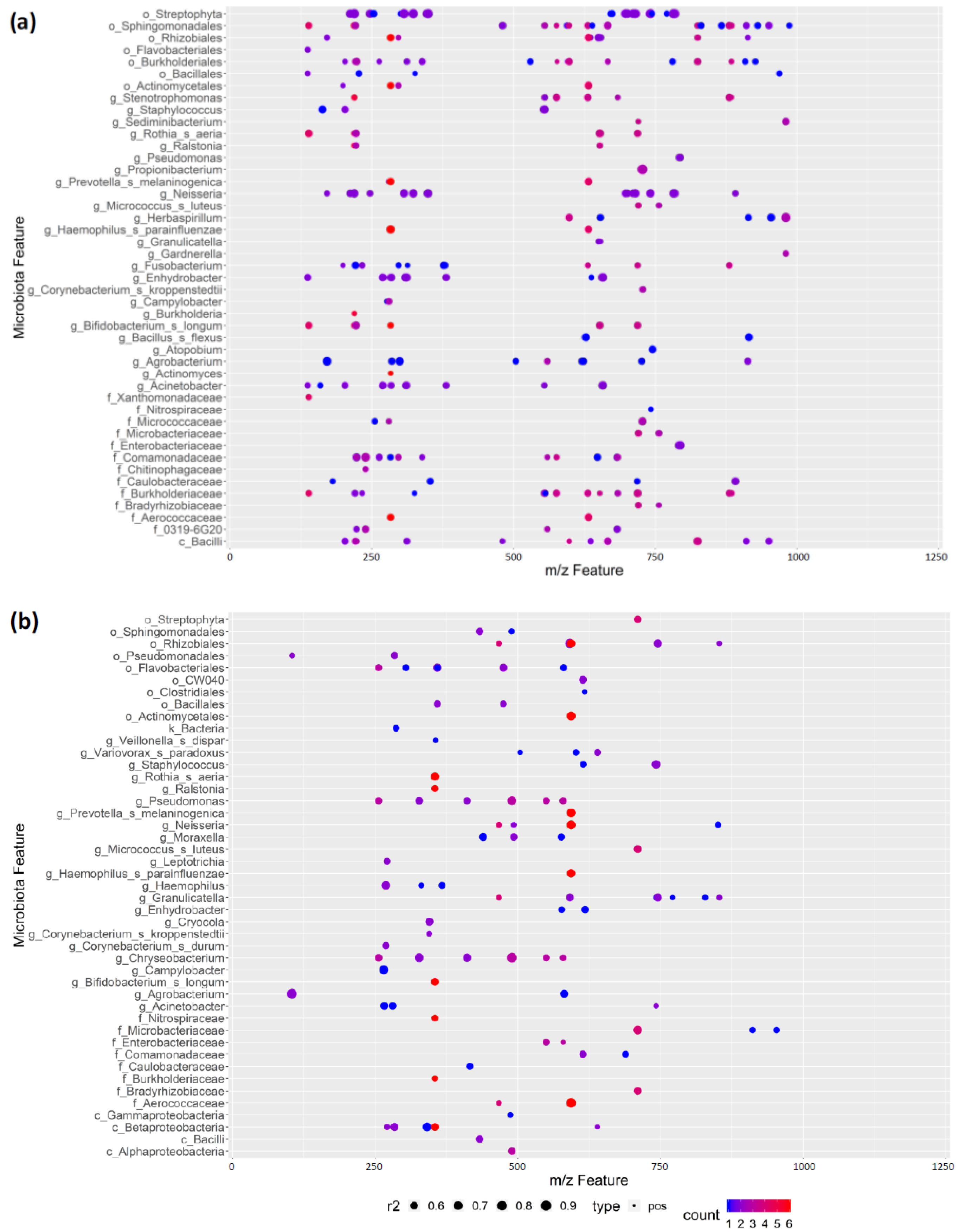
3.7. REIMS Shows Potential Co-Occurrence of Microbiota and Metabolite Features
3.8. Implications for Human Milk Banking
4. Limitations
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Piantadosi, S.T.; Kidd, C. Extraordinary intelligence and the care of infants. Proc. Natl. Acad. Sci. USA 2016, 113, 6874–6879. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oftedal, O.T. The evolution of milk secretion and its ancient origins. Animal 2012, 6, 355–368. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andreas, N.J.; Kampmann, B.; Mehring Le-Doare, K. Human breast milk: A review on its composition and bioactivity. Early Hum. Dev. 2015, 91, 629–635. [Google Scholar] [CrossRef] [PubMed]
- Blesa, M.; Sullivan, G.; Anblagan, D.; Telford, E.J.; Quigley, A.J.; Sparrow, S.A.; Serag, A.; Semple, S.I.; Bastin, M.E.; Boardman, J.P. Early breast milk exposure modifies brain connectivity in preterm infants. Neuroimage 2019, 184, 431–439. [Google Scholar] [CrossRef] [PubMed]
- Lewandowski, A.J.; Lamata, P.; Francis, J.M.; Piechnik, S.K.; Ferreira, V.M.; Boardman, H.; Neubauer, S.; Singhal, A.; Leeson, P.; Lucas, A. Breast Milk Consumption in Preterm Neonates and Cardiac Shape in Adulthood. Pediatrics 2016, 138, e20160050. [Google Scholar] [CrossRef] [Green Version]
- Bode, L. The functional biology of human milk oligosaccharides. Early Hum. Dev. 2015, 91, 619–622. [Google Scholar] [CrossRef]
- Forbes, J.D.; Azad, M.B.; Vehling, L.; Tun, H.M.; Konya, T.B.; Guttman, D.S.; Field, C.J.; Lefebvre, D.; Sears, M.R.; Becker, A.B.; et al. Association of Exposure to Formula in the Hospital and Subsequent Infant Feeding Practices with Gut Microbiota and Risk of Overweight in the First Year of Life. JAMA Pediatr. 2018, 172, e181161. [Google Scholar] [CrossRef]
- Moossavi, S.; Azad, M.B. Quantifying and Interpreting the Association between Early-Life Gut Microbiota Composition and Childhood Obesity. mBio 2019, 10, e02787-18. [Google Scholar] [CrossRef] [Green Version]
- Stewart, C.J.; Ajami, N.J.; O’Brien, J.L.; Hutchinson, D.S.; Smith, D.P.; Wong, M.C.; Ross, M.C.; Lloyd, R.E.; Doddapaneni, H.; Metcalf, G.A.; et al. Temporal development of the gut microbiome in early childhood from the TEDDY study. Nature 2018, 562, 583–588. [Google Scholar] [CrossRef]
- Vatanen, T.; Franzosa, E.A.; Schwager, R.; Tripathi, S.; Arthur, T.D.; Vehik, K.; Lernmark, Å.; Hagopian, W.A.; Rewers, M.J.; She, J.X.; et al. The human gut microbiome in early-onset type 1 diabetes from the TEDDY study. Nature 2018, 562, 589–594. [Google Scholar] [CrossRef]
- Goodwin, G. Type 1 Diabetes Mellitus and Celiac Disease: Distinct Autoimmune Disorders That Share Common Pathogenic Mechanisms. Horm. Res. Paediatr. 2019, 92, 285–292. [Google Scholar] [CrossRef] [PubMed]
- Johnson, R.K.; Vanderlinden, L.; DeFelice, B.C.; Kechris, K.; Uusitalo, U.; Fiehn, O.; Sontag, M.; Crume, T.; Beyerlein, A.; Lernmark, Å.; et al. Metabolite-related dietary patterns and the development of islet autoimmunity. Sci. Rep. 2019, 9, 14819. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dettwyler, K.A. Duration of breast feeding and adult arterial distensibility. Humans are primates, designed to breast feed for years not months. BMJ 2001, 323, 691–693. [Google Scholar] [CrossRef]
- Goldman, A.S. Evolution of the mammary gland defense system and the ontogeny of the immune system. J. Mammary Gland Biol. Neoplasia 2002, 7, 277–289. [Google Scholar] [CrossRef]
- Perrin, M.T.; Fogleman, A.D.; Newburg, D.S.; Allen, J.C. A longitudinal study of human milk composition in the second year postpartum: Implications for human milk banking. Matern. Child Nutr. 2017, 13, e12239. [Google Scholar] [CrossRef] [Green Version]
- Czosnykowska-Łukacka, M.; Królak-Olejnik, B.; Orczyk-Pawiłowicz, M. Breast Milk Macronutrient Components in Prolonged Lactation. Nutrients 2018, 10, 1893. [Google Scholar] [CrossRef] [Green Version]
- Garwolińska, D.; Hewelt-Belka, W.; Namieśnik, J.; Kot-Wasik, A. Rapid Characterization of the Human Breast Milk Lipidome Using a Solid-Phase Microextraction and Liquid Chromatography-Mass Spectrometry-Based Approach. J. Proteome Res. 2017, 16, 3200–3208. [Google Scholar] [CrossRef]
- Beck, K.L.; Weber, D.; Phinney, B.S.; Smilowitz, J.T.; Hinde, K.; Lönnerdal, B.; Korf, I.; Lemay, D.G. Comparative proteomics of human and macaque milk reveals species-specific nutrition during postnatal development. J. Proteome Res. 2015, 14, 2143–2157. [Google Scholar] [CrossRef]
- Hunt, K.M.; Foster, J.A.; Forney, L.J.; Schütte, U.M.; Beck, D.L.; Abdo, Z.; Fox, L.K.; Williams, J.E.; McGuire, M.K.; McGuire, M.A. Characterization of the diversity and temporal stability of bacterial communities in human milk. PLoS ONE 2011, 6, e21313. [Google Scholar] [CrossRef] [Green Version]
- Lässer, C.; Alikhani, V.S.; Ekström, K.; Eldh, M.; Paredes, P.T.; Bossios, A.; Sjöstrand, M.; Gabrielsson, S.; Lötvall, J.; Valadi, H. Human saliva, plasma and breast milk exosomes contain RNA: Uptake by macrophages. J. Transl. Med. 2011, 9, 9. [Google Scholar] [CrossRef] [Green Version]
- Sundekilde, U.; Downey, E.; O’Mahony, J.; O’Shea, C.-A.; Ryan, C.; Kelly, A.; Bertram, H. The effect of gestational and lactational age on the human milk metabolome. Nutrients 2016, 8, 304. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cameron, S.J.; Takáts, Z. Mass Spectrometry Approaches to Metabolic Profiling of Microbial Communities within the Human Gastrointestinal Tract. Methods 2018, 149, 13–24. [Google Scholar] [CrossRef] [PubMed]
- Cameron, S.J.S.; Huws, S.; Hegarty, M.J.; Smith, D.P.M.; Mur, L.A.J. The Human Salivary Microbiome Exhibits Temporal Stability in Bacterial Diversity. FEMS Microbiol. Ecol. 2015, 91, fiv091. [Google Scholar] [CrossRef] [PubMed]
- Mullish, B.H.; Pechlivanis, A.; Barker, G.F.; Thursz, M.R.; Marchesi, J.R.; McDonald, J.A. Functional microbiomics: Evaluation of gut microbiota-bile acid metabolism interactions in health and disease. Methods 2018, 149, 49–58. [Google Scholar] [CrossRef] [Green Version]
- Bolt, F.; Cameron, S.J.; Karancsi, T.; Simon, D.; Schaffer, R.; Rickards, T.; Hardiman, K.; Burke, A.; Bodai, Z.; Perdones-Montero, A.; et al. Automated High-Throughput Identification and Characterization of Clinically Important Bacteria and Fungi using Rapid Evaporative Ionization Mass Spectrometry. Anal. Chem. 2016, 88, 9419–9426. [Google Scholar] [CrossRef]
- Cameron, S.J.; Bodai, Z.; Temelkuran, B.; Perdones-Montero, A.; Bolt, F.; Burke, A.; Alexander-Hardiman, K.; Salzet, M.; Fournier, I.; Rebec, M.; et al. Utilisation of Ambient Laser Desorption Ionisation Mass Spectrometry (ALDI-MS) Improves Lipid-Based Microbial Species Level Identification. Sci. Rep. 2019, 9, 3006. [Google Scholar] [CrossRef] [Green Version]
- Mitoulas, L.R.; Lai, C.T.; Gurrin, L.C.; Larsson, M.; Hartmann, P.E. Efficacy of breast milk expression using an electric breast pump. J. Hum. Lact. 2002, 18, 344–352. [Google Scholar] [CrossRef]
- Salter, S.J.; Cox, M.J.; Turek, E.M.; Calus, S.T.; Cookson, W.O.; Moffatt, M.F.; Turner, P.; Parkhill, J.; Loman, N.J.; Walker, A.W. Reagent and Laboratory Contamination can Critically Impact Sequence-Based Microbiome Analyses. BMC Biol. 2014, 12, 87. [Google Scholar] [CrossRef] [Green Version]
- Douglas, C.A.; Ivey, K.L.; Papanicolas, L.E.; Best, K.P.; Muhlhausler, B.S.; Rogers, G.B. DNA extraction approaches substantially influence the assessment of the human breast milk microbiome. Sci. Rep. 2020, 10, 123. [Google Scholar] [CrossRef]
- Moossavi, S.; Sepehri, S.; Robertson, B.; Bode, L.; Goruk, S.; Field, C.J.; Lix, L.M.; de Souza, R.J.; Becker, A.B.; Mandhane, P.J. Composition and variation of the human milk microbiota are influenced by maternal and early-life factors. Cell Host Microbe 2019, 25, 324–335.e324. [Google Scholar] [CrossRef] [Green Version]
- Cabrera-Rubio, R.; Collado, M.C.; Laitinen, K.; Salminen, S.; Isolauri, E.; Mira, A. The Human Milk Microbiome Changes over Lactation and is Shaped by Maternal Weight and Mode of Delivery. Am. J. Clin. Nutr. 2012, 96, 544–551. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fernández, L.; Langa, S.; Martín, V.; Maldonado, A.; Jiménez, E.; Martín, R.; Rodríguez, J.M. The human milk microbiota: Origin and potential roles in health and disease. Pharmacol. Res. 2013, 69, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Sundekilde, U.; Larsen, L.; Bertram, H. NMR-based milk metabolomics. Metabolites 2013, 3, 204–222. [Google Scholar] [CrossRef] [PubMed]
- Perrone, S.; Longini, M.; Zollino, I.; Bazzini, F.; Tassini, M.; Vivi, A.; Bracciali, C.; Calderisi, M.; Buonocore, G. Breast milk: To each his own. From metabolomic study, evidence of personalized nutrition in preterm infants. Nutrition 2019, 62, 158–161. [Google Scholar] [CrossRef] [PubMed]
- Marincola, F.C.; Noto, A.; Caboni, P.; Reali, A.; Barberini, L.; Lussu, M.; Murgia, F.; Santoru, M.L.; Atzori, L.; Fanos, V. A metabolomic study of preterm human and formula milk by high resolution NMR and GC/MS analysis: Preliminary results. J. Matern.-Fetal Neonatal Med. 2012, 25, 62–67. [Google Scholar] [CrossRef] [PubMed]
- WHO. Global Strategy for Infant and Young Child Feeding; WHO: Geneva, Switzerland, 2010; Available online: https://www.who.int/entity/nutrition/topics/global_strategy/en/index.html (accessed on 2 October 2020).
- Mandel, D.; Lubetzky, R.; Dollberg, S.; Barak, S.; Mimouni, F.B. Fat and energy contents of expressed human breast milk in prolonged lactation. Pediatrics 2005, 116, e432–e435. [Google Scholar] [CrossRef] [Green Version]
- Verd, S.; Ginovart, G.; Calvo, J.; Ponce-Taylor, J.; Gaya, A. Variation in the protein composition of human milk during extended lactation: A narrative review. Nutrients 2018, 10, 1124. [Google Scholar] [CrossRef] [Green Version]
- Brown, S.L.; Bright, R.A.; Dwyer, D.E.; Foxman, B. Breast pump adverse events: Reports to the Food and Drug Administration. J. Hum. Lact. 2005, 21, 169–174. [Google Scholar] [CrossRef]
- Boo, N.-Y.; Nordiah, A.; Alfizah, H.; Nor-Rohaini, A.; Lim, V. Contamination of breast milk obtained by manual expression and breast pumps in mothers of very low birthweight infants. J. Hosp. Infect. 2001, 49, 274–281. [Google Scholar] [CrossRef]
- Quinn, L.; Ailsworth, M.; Matthews, E.; Kellams, A.; Shirley, D.-A. Serratia marcescens colonization causing pink breast milk and pink diapers: A case report and literature review. Breastfeed Med. 2018, 13, 388–394. [Google Scholar] [CrossRef]
- Flores-Antón, B.; Martín-Cornejo, J.; Morante-Santana, M.; García-Lara, N.; Sierra-Colomina, G.; De la Cruz-Bértolo, J.; Martín-Arriscado-Arroba, C.; Escuder-Vieco, D.; Soriano-Ramos, M.; Chaves, F. Comparison of two methods for cleaning breast pump milk collection kits in human milk banks. J. Hosp. Infect. 2019, 103, 217–222. [Google Scholar] [CrossRef] [PubMed]
- Innis, S.M. Impact of maternal diet on human milk composition and neurological development of infants. Am. J. Clin. Nutr. 2014, 99, 734S–741S. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Olafsdottir, A.S.; Wagner, K.-H.; Thorsdottir, I.; Elmadfa, I. Fat-soluble vitamins in the maternal diet, influence of cod liver oil supplementation and impact of the maternal diet on human milk composition. Ann. Nutr. Metab. 2001, 45, 265–272. [Google Scholar] [CrossRef] [PubMed]
- Lyons, K.E.; Ryan, C.A.; Dempsey, E.M.; Ross, R.P.; Stanton, C. Breast Milk, a Source of Beneficial Microbes and Associated Benefits for Infant Health. Nutrients 2020, 12, 1039. [Google Scholar] [CrossRef] [PubMed]
- Gopalakrishna, K.P.; Hand, T.W. Influence of Maternal Milk on the Neonatal Intestinal Microbiome. Nutrients 2020, 12, 823. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sela, D.A.; Mills, D.A. Nursing our microbiota: Molecular linkages between bifidobacteria and milk oligosaccharides. Trends Microbiol. 2010, 18, 298–307. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hakkarainen, J.; Toivanen, M.; Leinonen, A.; Frängsmyr, L.; Strömberg, N.; Lapinjoki, S.; Nassif, X.; Tikkanen-Kaukanen, C. Human and bovine milk oligosaccharides inhibit Neisseria meningitidis pili attachment in vitro. J. Nutr. 2005, 135, 2445–2448. [Google Scholar] [CrossRef] [Green Version]
- PATH. Policy Brief: Ensuring equitable access to human milk for all infants: A comprehensive approach to essential newborn care. In Global Breastfeeding Collective; PATH: Seattle, WA, USA, 2017; Available online: https://path.azureedge.net/media/documents/MNCHN_EquitableAccesstoHumanMilk_PolicyBrief.pdf (accessed on 2 October 2020).
- Khan, S.; Hepworth, A.R.; Prime, D.K.; Lai, C.T.; Trengove, N.J.; Hartmann, P.E. Variation in fat, lactose, and protein composition in breast milk over 24 hours: Associations with infant feeding patterns. J. Hum. Lact. 2013, 29, 81–89. [Google Scholar] [CrossRef]
- NICE. Donor Milk Banks: The Operation of Donor Milk Bank Services. Available online: http://www.nice.org.uk/guidance/cg93 (accessed on 26 May 2020).
- Triantis, V.; Bode, L.; van Neerven, R.J.J. Immunological Effects of Human Milk Oligosaccharides. Front. Pediatr. 2018, 6, 190. [Google Scholar] [CrossRef]
- Lin, A.E.; Autran, C.A.; Szyszka, A.; Escajadillo, T.; Huang, M.; Godula, K.; Prudden, A.R.; Boons, G.J.; Lewis, A.L.; Doran, K.S.; et al. Human milk oligosaccharides inhibit growth of group B. J. Biol. Chem. 2017, 292, 11243–11249. [Google Scholar] [CrossRef] [Green Version]
- Bode, L. Human milk oligosaccharides: Every baby needs a sugar mama. Glycobiology 2012, 22, 1147–1162. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Selkrig, J.; Wong, P.; Zhang, X.; Pettersson, S. Metabolic tinkering by the gut microbiome: Implications for brain development and function. Gut Microbes 2014, 5, 369–380. [Google Scholar] [CrossRef] [PubMed]
- Keunen, K.; van Elburg, R.M.; van Bel, F.; Benders, M.J. Impact of nutrition on brain development and its neuroprotective implications following preterm birth. Pediatr. Res. 2015, 77, 148–155. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Victora, C.G.; Bahl, R.; Barros, A.J.; França, G.V.; Horton, S.; Krasevec, J.; Murch, S.; Sankar, M.J.; Walker, N.; Rollins, N.C.; et al. Breastfeeding in the 21st century: Epidemiology, mechanisms, and lifelong effect. Lancet 2016, 387, 475–490. [Google Scholar] [CrossRef] [Green Version]
- Greaves, M. A causal mechanism for childhood acute lymphoblastic leukaemia. Nat. Rev. Cancer 2018, 18, 471–484. [Google Scholar] [CrossRef]
- Klopp, A.; Vehling, L.; Becker, A.B.; Subbarao, P.; Mandhane, P.J.; Turvey, S.E.; Lefebvre, D.L.; Sears, M.R.; Azad, M.B.; Investigators, C.S. Modes of Infant Feeding and the Risk of Childhood Asthma: A Prospective Birth Cohort Study. J. Pediatr. 2017, 190, 192–199.e192. [Google Scholar] [CrossRef] [PubMed]


| Age Group | Expression Method | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 03 to 06 | 06 to 12 | 12 to 24 | 24 to 36 | 36 to 48 | p Value | H | M | E | p Value | |
| Total Participants | 12 | 12 | 16 | 14 | 8 | N/A | 7 | 10 | 45 | N/A |
| Age (Years) | 33.75 (3.41) | 34.75 (2.80) | 32.81 (4.90) | 35.29 (5.55) | 37.63 (5.76) | 0.24 ‡ | 35.86 (4.53) | 33.60 (5.68) | 34.56 (4.56) | 0.77 ‡ |
| Ethnicity | ||||||||||
| Caribbean | 0 | 0 | 0 | 1 | 1 | N/A * | 0 | 1 | 1 | N/A * |
| Chinese | 0 | 0 | 1 | 0 | 1 | N/A * | 0 | 0 | 2 | N/A * |
| Other | 1 | 0 | 0 | 0 | 0 | N/A * | 0 | 1 | 0 | N/A * |
| White | 11 | 12 | 12 | 12 | 5 | N/A * | 6 | 8 | 38 | N/A * |
| White Irish | 0 | 0 | 1 | 0 | 0 | N/A * | 0 | 0 | 1 | N/A * |
| White Mixed | 0 | 0 | 1 | 1 | 1 | N/A * | 1 | 0 | 2 | N/A * |
| White Other | 0 | 0 | 1 | 0 | 0 | N/A * | 0 | 0 | 1 | N/A * |
| Body Mass Index | ||||||||||
| Pre-Pregnancy | 24.57 (4.99) | 27.21 (4.83) | 23.95 (4.22) | 26.14 (4.30) | 24.21 (4.24) | 0.32 † | 25.96 (4.86) | 26.09 (4.33) | 24.92 (4.61) | 0.70 † |
| Post-Pregnancy | 25.79 (4.60) | 27.82 (4.82) | 25.01 (3.92) | 26.98 (5.42) | 26.27 (5.13 | 0.59 † | 26.34 (5.68) | 26.66 (3.87) | 26.23 (4.81) | 0.97 † |
| Previous Pregnancies | 1.58 (0.79) | 1.50 (0.52) | 1.38 (0.62) | 1.57 (0.93) | 1.50 (0.93) | 0.95 ‡ | 1.57 (0.98) | 1.50 (0.53) | 1.49 (0.73) | 0.95 ‡ |
| Sex of Nursling | ||||||||||
| Female | 6 | 8 | 8 | 5 | 3 | 0.57 * | 2 | 6 | 22 | 0.44 * |
| Male | 6 | 4 | 8 | 9 | 5 | 5 | 4 | 23 | ||
| Diet | ||||||||||
| Meat Eater | 11 | 10 | 16 | 9 | 6 | N/A * | 6 | 8 | 38 | N/A * |
| Pescetarian | 1 | 0 | 0 | 2 | 1 | 0 | 1 | 3 | ||
| Vegetarian | 0 | 2 | 0 | 3 | 1 | 1 | 1 | 4 | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shenker, N.S.; Perdones-Montero, A.; Burke, A.; Stickland, S.; McDonald, J.A.K.; Alexander-Hardiman, K.; Flanagan, J.; Takats, Z.; Cameron, S.J.S. Metabolomic and Metataxonomic Fingerprinting of Human Milk Suggests Compositional Stability over a Natural Term of Breastfeeding to 24 Months. Nutrients 2020, 12, 3450. https://0-doi-org.brum.beds.ac.uk/10.3390/nu12113450
Shenker NS, Perdones-Montero A, Burke A, Stickland S, McDonald JAK, Alexander-Hardiman K, Flanagan J, Takats Z, Cameron SJS. Metabolomic and Metataxonomic Fingerprinting of Human Milk Suggests Compositional Stability over a Natural Term of Breastfeeding to 24 Months. Nutrients. 2020; 12(11):3450. https://0-doi-org.brum.beds.ac.uk/10.3390/nu12113450
Chicago/Turabian StyleShenker, Natalie S., Alvaro Perdones-Montero, Adam Burke, Sarah Stickland, Julie A.K. McDonald, Kate Alexander-Hardiman, James Flanagan, Zoltan Takats, and Simon J.S. Cameron. 2020. "Metabolomic and Metataxonomic Fingerprinting of Human Milk Suggests Compositional Stability over a Natural Term of Breastfeeding to 24 Months" Nutrients 12, no. 11: 3450. https://0-doi-org.brum.beds.ac.uk/10.3390/nu12113450





