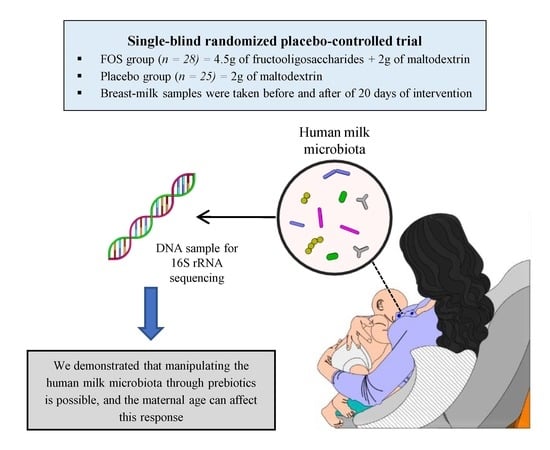
Response of the Human Milk Microbiota to a Maternal Prebiotic Intervention Is Individual and Influenced by Maternal Age
Abstract
:1. Introduction
2. Materials and Methods
2.1. Subject and Study Design
2.2. Data Collection
2.3. Milk Samples Collection
2.4. DNA Isolation
2.5. PCR Amplification for Sequencing
2.6. Sequence Processing
2.7. Data Deposition and Materials Sharing
2.8. Quantitative PCR
2.9. Statistical Analysis
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Clemente, J.C.; Ursell, L.K.; Parfrey, L.W.; Knight, R. The impact of the gut microbiota on human health: An integrative view. Cell 2012, 148, 1258–1270. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Asnicar, F.; Manara, S.; Zolfo, M.; Truong, D.T.; Scholz, M.; Armanini, F.; Ferreti, P.; Gorfer, V.; Pedrotti, A.; Tett, A.; et al. Studying vertical microbiome transmission from mothers to infants by strain-level metagenomic profiling. mSystems 2017, 2, e00164-16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johansson, M.A.; Sjögren, Y.M.; Persson, J.O.; Nilsson, C.; Sverremark-Ekström, E. Early colonization with a group of Lactobacilli decreases the risk for allergy at five years of age despite allergic heredity. PLoS ONE 2011, 6, e23031. [Google Scholar] [CrossRef] [PubMed]
- Algert, C.S.; Mcelduff, A.; Morris, J.M.; Roberts, C.L. Perinatal risk factors for early onset of Type 1 diabetes in a 2000–2005 birth cohort. Diabet. Med. 2009, 26, 1193–1197. [Google Scholar] [CrossRef] [Green Version]
- Kalliomäki, M.; Collado, M.C.; Salminen, S.; Isolauri, E. Early differences in fecal microbiota composition in children may predict overweight. Am. J. Clin. Nutr. 2008, 87, 534–538. [Google Scholar] [CrossRef]
- Ajslev, T.A.; Andersen, C.S.; Gamborg, M.; Sørensen, T.I.; Jess, T. Childhood overweight after establishment of the gut microbiota: The role of delivery mode, pre-pregnancy weight and early administration of antibiotics. Int. J. Obes. 2011, 35, 522–529. [Google Scholar] [CrossRef] [Green Version]
- Shen, T.C.D. Diet and gut microbiota in Health and disease. In Intestinal Microbiome: Functional Aspects in Health and Disease; Isolauri, E., Sherman, P.M., Walker, W.A., Eds.; Karger Publishers: Basel, Switzerland, 2017; pp. 117–126. [Google Scholar]
- Korpela, K.; De Vos, W. Early life colonization of the human gut: Microbes matter everywhere. Curr. Opin. Microbiol. 2019, 44, 70–78. [Google Scholar] [CrossRef] [Green Version]
- Wen, L.; Duffy, A. Factors influencing the gut microbiota, inflammation, and type 2 Diabetes. J. Nutr. 2017, 147, 1468S–1475S. [Google Scholar] [CrossRef] [Green Version]
- Funkhouser, L.J.; Bordenstein, S.R. Mom Knows Best: The Universality of Maternal Microbial Transmission. PLoS Biol. 2013, 11, e1001631. [Google Scholar] [CrossRef]
- Bäckhed, F.; Roswall, J.; Peng, Y.; Feng, Q.; Jia, H.; Kovatcheva-Datchary, P.; Li, Y.; Xia, Y.; Xie, H.; Zhong, H.; et al. Dynamics and stabilization of the human gut microbiome during the first year of life. Cell Host Microbe 2015, 17, 690–703. [Google Scholar] [CrossRef] [Green Version]
- Gregory, K.E.; Samuel, B.S.; Houghteling, P.; Shan, G.; Ausubel, F.M.; Sadreyev, R.I.; Walker, W.A. Influence of maternal breast milk ingestion on acquisition of the intestinal microbiome in preterm infants. Microbiome 2016, 4, 68. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carlson, J.; Slavin, J. Health benefits of fiber, prebiotics and probiotics: A review of intestinal health and related health claims. Qual. Assur. Saf. Crop. 2016, 8, 539–553. [Google Scholar] [CrossRef]
- Xavier-Santos, D.; Bedani, R.; Lima, E.D.; Saad, S.M.I. Impact of probiotics and prebiotics targeting metabolic syndrome. J. Funct. Foods 2020, 64, 103666. [Google Scholar] [CrossRef]
- Gibson, G.R.; Hutkins, R.; Sanders, M.E.; Prescott, S.L.; Reimer, R.A.; Salminen, S.; Scott, K.; Stanton, C.; Swanson, K.; Cani, P.D.; et al. The International Scientific Association for Probiotics and Prebiotics (ISAPP) consensus statement on the definition and scope of prebiotics. Nat. Rev. Gastro. Hepat. 2017, 14, 491–502. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Champ, M.; Hoebler, C. Functional food for pregnant, lactating women and in perinatal nutrition: A role for dietary fibers? Curr. Opin. Clin. Nutr. Metab. Care 2009, 12, 565–574. [Google Scholar] [CrossRef] [PubMed]
- Kolida, S.; Gibson, G.R. Synbiotics in health and disease. Annu. Rev. Food Sci. Technol. 2011, 2, 373–393. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Charbonneau, M.R.; O’donnell, D.; Blanton, L.V.; Totten, S.M.; Davis, J.C.; Barratt, M.J.; Cheng, J.; Guruge, J.; Talcott, M.; Bain, J.R.; et al. Sialylated milk oligosaccharides promote microbiota-dependent growth in models of infant undernutrition. Cell 2016, 164, 859–871. [Google Scholar] [CrossRef]
- Milani, C.; Duranti, S.; Bottacini, F.; Casey, E.; Turronia, F.; Mahony, J.; Belzer, C.; Palacio, S.D.; Montes, S.A.; Mancabelli, L.; et al. The first microbial colonizers of the human gut: Composition, activities, and health implications of the infant gut microbiota. Microbiol. Mol. Biol. R. 2017, 81, 1–67. [Google Scholar] [CrossRef] [Green Version]
- Martín, R.; Jiménez, E.; Heilig, G.H.J.; Fernández, L.; Marín, M.L.; Zoetendal, E.G.; Rodríguez, J.M. Isolation of bifidobacteria from breast milk and assessment of the bifidobacterial population by PCR-denaturing gradient gel electrophoresis and quantitative real-time PCR. Appl. Environ. Microbiol. 2009, 75, 965–969. [Google Scholar] [CrossRef] [Green Version]
- Jost, T.; Lacroix, C.; Braegger, C.; Chassard, C. Assessment of bacterial diversity in breast milk using culture-dependent and culture-independent approaches. Br. J. Nutr. 2013, 14, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Yu, Y.; Lee, C.; Kim, J.; Hwang, S. Group-Specific primer and probe sets to detect methanogenic communities using quantitative real-time polymerase chain reaction. Biotechnol. Bioeng. 2005, 89, 670–679. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paulson, J.; Stine, O.; Bravo, H.; Pop, M. Differential Abundance Analysis for Microbial Marker-Gene Surveys. Nat. Methods 2013, 10, 1200–1202. [Google Scholar] [CrossRef] [Green Version]
- Mcmurdie, P.; Holmes, S. Waste Not, Want Not: Why Rarefying Microbiome Data Is Inadmissible. PLoS Comput. Biol. 2014, 8, e61217. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oksanen, J.; Blanchet, F.G.; Friendly, M.; Kindt, R.; Legendre, P.; Mcglinn, D.; Minchin, P.R.; O’hara, R.B.; Simpson, G.L.; Solymos, P.; et al. Vegan: Community Ecology Package. R Package Vegan, Vers2.2-1. 2016. Available online: http://outputs.worldagroforestry.org/cgi-bin/koha/opac-detail.pl?biblionumber=39154 (accessed on 17 September 2019).
- Wickham, H. Ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2009. [Google Scholar]
- Furet, J.P.; Firmesse, O.; Gourmelon, M.; Bridonneau, C.; Tap, J.; Mondot, S.; Doré, J.; Corthier, G. Comparative assessment of human and farm animal faecal microbiota using real-time quantitative PCR. FEMS Microbiol. Ecol. 2009, 68, 351–362. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rinttilä, T.; Kassinen, A.; Malinen, E.; Krogius, L.; Palva, A. Development of an extensive set of 16S rDNA-targeted primers for quantification of pathogenic and indigenous bacteria in faecal samples by real-time PCR. J. Appl. Microbiol. 2004, 97, 1166–1177. [Google Scholar] [CrossRef]
- Mandal, S.; van Treuren, W.; White, R.A.; Eggesbø, M.; Knight, R.; Peddada, S.D. Analysis of composition of microbiomes: A novel method for studying microbial composition. Microb. Ecol. Health Dis. 2015, 26, 27663. [Google Scholar] [CrossRef] [Green Version]
- Yasmin, A.; Butt, M.S.; Afzaal, M.; Van Baak, M.; Nadeem, M.T.; Shahid, M.Z. Prebiotics, gut microbiota and metabolic risks: Unveiling the relationship. J. Funct. Foods 2015, 17, 189–201. [Google Scholar] [CrossRef]
- Jost, T.; Lacroix, C.; Braegger, C.; Chassard, C. Stability of the maternal gut microbiota during late pregnancy and early lactation. Curr. Microbiol. 2014, 68, 419–427. [Google Scholar] [CrossRef] [Green Version]
- Martín, V.; Maldonado-Barragán, A.; Moles, L.; Rodriguez-Baños, M.; Del Campo, R.; Fernández, L.; Rodríguez, J.M.; Jiménez, E. Sharing of bacterial strains between breast milk and infant feces. J. Hum. Lact. 2012, 28, 36–44. [Google Scholar] [CrossRef] [PubMed]
- Pokusaeva, K.; Fitzgerald, G.F.; van Sinderen, D. Carbohydrate metabolism in Bifidobacteria. Genes Nutr. 2011, 6, 285–306. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shadid, R.; Haarman, M.; Knol, J.; Theis, W.; Beermann, C.; Ejosk-Dendorfer, D.; Schendel, D.J.; Koletzko, B.V.; Krauss-Etschmann, S. Effects of galactooligosaccharide and long-chain fructooligosaccharide supplementation during pregnancy on maternal and neonatal microbiota and immunity: A randomized, double-blind, placebo-controlled study. Am. J. Clin. Nutr. 2007, 86, 1426–1437. [Google Scholar] [CrossRef] [PubMed]
- Padilha, M.; Danneskiold-Samsøe, N.B.; Brejnrod, A.; Hoffmann, C.; Cabral, V.P.; Iaucci, J.M.; Sales, C.H.; Fisberg, R.M.; Cortez, R.V.; Brix, S.; et al. The Human Milk Microbiota is Modulated by Maternal Diet. Microorganisms 2019, 7, 502. [Google Scholar] [CrossRef] [Green Version]
- Odamaki, T.; Kato, K.; Sugahara, H.; Hashikura, N.; Takahashi, S.; Xiao, J.Z.; Abe, F.; Osawa, R. Age-related changes in gut microbiota composition from newborn to centenarian: A cross-sectional study. BMC Microbiol. 2016, 16, 90. [Google Scholar] [CrossRef] [Green Version]
- Parsons, T.J.; Manor, O.; Power, C. Changes in diet and physical activity in the 1990s in a large British sample (1958 birth cohort). Eur. J. Clin. Nutr. 2005, 59, 49–56. [Google Scholar] [CrossRef] [PubMed]
- Mishra, G.; McNaughton, S.; Bramwell, G.; Wadsworth, M. Longitudinal changes in dietary patterns during adult life. Br. J. Nutr. 2006, 96, 735–744. [Google Scholar]
- Wu, G.D.; Chen, J.; Hoffmann, C.; Bittinger, K.; Chen, Y.Y.; Keilbaugh, S.A.; Bewtra, M.; Knights, D.; Walters, W.A.; Knight, R.; et al. Linking long-term dietary patterns with gut microbial enterotypes. Science 2011, 334, 105–108. [Google Scholar] [CrossRef] [Green Version]
- Walker, A.W.; Sanderson, J.D.; Churcher, C.; Parkes, G.C.; Hudspith, B.N.; Rayment, N.; Brostoff, J.; Parkhill, J.; Dougan, G.; Petrovska, L. High-throughput clone library analysis of the mucosa-associated microbiota reveals dysbiosis and differences between inflamed and non-inflamed regions of the intestine in inflammatory bowel disease. BMC Microbiol. 2011, 11, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Griffin, N.W.; Ahern, P.P.; Cheng, J.; Heath, A.C.; Ilkayeva, O.; Newgard, C.B.; Fontana, L.; Gordon, J.I. Prior dietary practices and connections to a human gut microbial metacommunity alter responses to diet interventions. Cell Host Microbe 2017, 21, 84–96. [Google Scholar] [CrossRef] [Green Version]
- Long, W.; Xue, Z.; Zhang, Q.; Feng, Z.; Bridgewater, L.; Wang, L.; Zhao, L.; Pang, X. Differential responses of gut microbiota to the same prebiotic formula in oligotrophic and eutrophic batch fermentation systems. Sci. Rep. 2015, 5, 13469. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martínez, I.; Lattimer, J.M.; Hubach, K.L.; Case, J.A.; Yang, J.; Weber, C.G.; Louk, J.A.; Rose, D.J.; Kyureghian, G.; Peterson, D.A.; et al. Gut microbiome composition is linked to whole grain-induced immunological improvements. ISME J 2013, 7, 269–280. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nikniaz, L.; Ostadrahimi, A.; Mahdavi, R.; Hejazi, M.A.; Salekdeh, G.H. Effects of synbiotic supplementation on breast milk levels of IgA, TGF-β1 and TGF-β2. J. Hum. Lact. 2013, 29, 591–596. [Google Scholar] [PubMed]
- Kubota, T.; Shimojo, N.; Nonaka, K.; Yamashita, M.; Ohara, O.; Igoshi, Y.; Ozawa, N.; Nakano, T.; Morita, Y.; Yuzaburo, I.; et al. Prebiotic consumption in pregnant and lactating women increases IL-27 expression in human milk. Br. J. Nutr. 2014, 111, 625–632. [Google Scholar]
- Collado, M.C.; Laitinen, K.; Salminen, S.; Isolauri, E. Maternal weight and excessive weight gain during pregnancy modify the immunomodulatory potential of breast milk. Pediatr. Res. 2012, 72, 77–85. [Google Scholar]




| Variable | Groups | ||
|---|---|---|---|
| FOS | Placebo | p-value ** | |
| n = 28 | n = 25 | ||
| Maternal Age (Years) | 28 (23–31) | 28 (21–31) | 0.469 |
| Race | 0.597 | ||
| Black/Brown | 12 (42.9) | 12 (48) | |
| White | 16 (57.1) | 13 (52) | |
| Family income estimated (USD/month) * | 770.4 (462.5–770.4) | 462.5 (462.5–770.39) | 0.458 |
| Duration of pregnancy (weeks) | 39 (38–40) | 39 (38–40) | 0.611 |
| Maternal antibiotic treatment | |||
| during pregnancy | 6 (21.4) | 7 (28) | 0.814 |
| during delivery | 11 (39.3) | 10 (40) | 1.000 |
| Alcohol drinking during pregnancy | 3 (10.7) | 1 (4) | 0.687 |
| Smoking during pregnancy | 2 (7.1) | 1 (4) | 1.000 |
| BMI before pregnancy (kg/m2) | 23.0 (20.7–25.2) | 22.6 (20.9–24.5) | 0.929 |
| Maternal weight gain over pregnancy (kg) | 11.2 (8–14.1) | 11.9 (8.3–16.6) | 0.497 |
| Anesthesia | 1.000 | ||
| No anesthesia | 7 (25) | 3 (12) | |
| Local | 13 (46.4) | 14 (56) | |
| Pudendal block | 0 | 0 | |
| Epidural | 0 | 4 (16) | |
| Spinal | 8 (28.6) | 3 (12) | |
| BMI at day 30 after delivery (before supplementation) | 23.6 (21.7–27.4) | 24.5 (22.3–26.9) | 0.879 |
| BMI at day 50 after delivery (after supplementation) | 24.1 (21.8–27.7) | 24.6 (22.1–26.5) | 0.973 |
| Baby gender | 0.940 | ||
| Male | 17 (60.7) | 14 (56) | |
| Female | 11 (39.3) | 11 (44) | |
| Birth weight (g) | 3205 (2933.7–3563.7) | 3210 (2977.5–3425) | 0.979 |
| Birth length (cm) | 48.5 (47.1–50) | 49 (47–50) | 1.000 |
| Infant diet before the supplementation | 0.145 | ||
| Exclusively breast milk | 23 (82.1) | 24 (96) | |
| Breast milk + formula | 5 (17.8) | 1 (4) | |
| Infant diet at day 50 after delivery after the supplementation | 0.145 | ||
| Exclusively breast milk | 21 (75) | 21 (84) | |
| Breast milk + formula | 7 (25) | 4 (16) | |
| Infant Δ weight over the intervention period (g) | 655.0 (458.7–832.5) | 792.5 (578.7–900.0) | 0.212 |
| Placebo | FOS | |||||
|---|---|---|---|---|---|---|
| Before | After | p-values * | Before | After | p-values * | |
| Energy (kcal) | 2091 (1950–2328) | 2295 (1956–2608) | 0.691 | 2266 (2105–2503) | 2161 (1960–2338) | 0.418 |
| Carbohydrates (g) | 299 (256–341) | 272 (257–319) | 0.731 | 355 (296–398) | 250 (227–338) | 0.006 |
| Proteins (g) | 76 (67–97) | 91 (68–110) | 0.560 | 80 (67–91) | 84 (74–98) | 0.866 |
| Fat (g) | 71 (55–83) | 72 (62–82) | 0.874 | 72 (63–81) | 76 (70–84) | 0.797 |
| Saturated fatty acids (g) | 23 (20–30) | 16 (11–18) | 0.691 | 26 (21–29) | 29 (21–31) | 0.797 |
| Monounsaturated fatty acids (g) | 20 (16–26) | 19 (17–26) | 0.560 | 22 (17–25) | 25 (21–29) | 0.884 |
| Polyunsaturated fatty acids (g) | 11 (9–13) | 28 (22–33) | 0.300 | 15 (13–18) | 16 (13–18) | 0.537 |
| Total dietary fibers (g) | 22 (19–23) | 22 (18–25) | 0.596 | 26 (21–35) | 22 (17–26) | 0.099 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Padilha, M.; Brejnrod, A.; Danneskiold-Samsøe, N.B.; Hoffmann, C.; Iaucci, J.d.M.; Cabral, V.P.; Xavier-Santos, D.; Taddei, C.R.; Kristiansen, K.; Saad, S.M.I. Response of the Human Milk Microbiota to a Maternal Prebiotic Intervention Is Individual and Influenced by Maternal Age. Nutrients 2020, 12, 1081. https://0-doi-org.brum.beds.ac.uk/10.3390/nu12041081
Padilha M, Brejnrod A, Danneskiold-Samsøe NB, Hoffmann C, Iaucci JdM, Cabral VP, Xavier-Santos D, Taddei CR, Kristiansen K, Saad SMI. Response of the Human Milk Microbiota to a Maternal Prebiotic Intervention Is Individual and Influenced by Maternal Age. Nutrients. 2020; 12(4):1081. https://0-doi-org.brum.beds.ac.uk/10.3390/nu12041081
Chicago/Turabian StylePadilha, Marina, Asker Brejnrod, Niels Banhos Danneskiold-Samsøe, Christian Hoffmann, Julia de Melo Iaucci, Vanessa Pereira Cabral, Douglas Xavier-Santos, Carla Romano Taddei, Karsten Kristiansen, and Susana Marta Isay Saad. 2020. "Response of the Human Milk Microbiota to a Maternal Prebiotic Intervention Is Individual and Influenced by Maternal Age" Nutrients 12, no. 4: 1081. https://0-doi-org.brum.beds.ac.uk/10.3390/nu12041081