Maternal Allergy and the Presence of Nonhuman Proteinaceous Molecules in Human Milk
Abstract
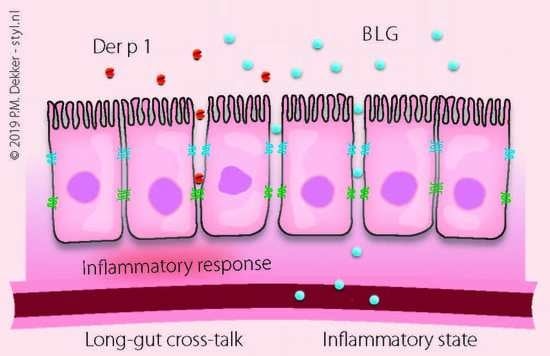
:1. Introduction
2. Materials and Methods
2.1. Milk Samples
2.2. Methods
2.2.1. Data Analysis
2.2.2. Annotation
2.2.3. Statistical Analysis
2.2.4. Confirmatory Analysis
3. Results
3.1. Identification of Exogenous Peptides
3.2. Differences between Allergic and Nonallergic Mothers
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Kilshaw, P.J.; Cant, A.J. The passage of maternal dietary proteins into human breast milk. Int. Arch. Allergy Appl. Immunol. 1984, 75, 8–15. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Garrigues, L.; Van Den Toorn, H.; Stahl, B.; Heck, A.J. Discovery and quantification of nonhuman proteins in human milk. J. Proteome Res. 2019, 18, 225–238. [Google Scholar] [CrossRef] [PubMed]
- Picariello, G.; Addeo, F.; Ferranti, P.; Nocerino, R.; Paparo, L.; Passariello, A.; Dallas, D.C.; Robinson, R.C.; Barile, D.; Canani, R.B. Antibody-independent identification of bovine milk-derived peptides in breast-milk. Food Funct. 2016, 7, 3402–3409. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Picariello, G.; De Cicco, M.; Nocerino, R.; Paparo, L.; Mamone, G.; Addeo, F.; Berni Canani, R. Excretion of dietary cow’s milk derived peptides into breast milk. Front. Nutr. 2019, 6, 25. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coscia, A.; Orrù, S.; Di Nicola, P.; Giuliani, F.; Varalda, A.; Peila, C.; Fabris, C.; Conti, A.; Bertino, E. Detection of cow’s milk proteins and minor components in human milk using proteomics techniques. J. Matern. Fetal Neonatal Med. 2012, 25, 49–51. [Google Scholar] [CrossRef] [PubMed]
- Schocker, F.; Baumert, J.; Kull, S.; Petersen, A.; Becker, W.M.; Jappe, U. Prospective investigation on the transfer of Ara h 2, the most potent peanut allergen, in human breast milk. Pediatr. Allergy Immunol. 2016, 27, 348–355. [Google Scholar] [CrossRef]
- Pastor-Vargas, C.; Maroto, A.S.; Diaz-Perales, A.; Villaba, M.; Casillas Diaz, N.; Vivanco, F.; Cuesta-Herranz, J. Sensitive detection of major food allergens in breast milk: First gateway for allergenic contact during breastfeeding. Allergy 2015, 70, 1024–1027. [Google Scholar] [CrossRef] [Green Version]
- Macchiaverni, P.; Rekima, A.; Turfkruyer, M.; Mascarell, L.; Airouche, S.; Moingeon, P.; Adel-Patient, K.; Condino-Neto, A.; Annesi-Maesano, I.; Prescott, S.L.; et al. Respiratory allergen from house dust mite is present in human milk and primes for allergic sensitization in a mouse model of asthma. Allergy 2014, 69, 395–398. [Google Scholar] [CrossRef]
- Vance, G.H.; Lewis, S.A.; Grimshaw, K.E.; Wood, P.J.; Briggs, R.A.; Thornton, C.A.; Warner, J.O. Exposure of the fetus and infant to hens’ egg ovalbumin via the placenta and breast milk in relation to maternal intake of dietary egg. Clin. Exp. Allergy 2005, 35, 1318–1326. [Google Scholar] [CrossRef]
- Reitsma, M.; Westerhout, J.; Wichers, H.J.; Wortelboer, H.M.; Verhoeckx, K.C. Protein transport across the small intestine in food allergy. Mol. Nutr. Food Res. 2014, 58, 194–205. [Google Scholar] [CrossRef]
- Benn, C.S.; Böttcher, M.F.; Pedersen, B.V.; Filteau, S.M.; Duchén, K. Mammary epithelial paracellular permeability in atopic and non-atopic mothers versus childhood atopy. Pediatr. Allergy Immunol. 2004, 15, 123–126. [Google Scholar] [CrossRef] [PubMed]
- Monks, J.; Neville, M.C. Albumin transcytosis across the epithelium of the lactating mouse mammary gland. J. Physiol. 2004, 560, 267–280. [Google Scholar] [CrossRef]
- Chirdo, F.G.; Rumbo, M.; Añón, M.C.; Fossati, C.A. Presence of high levels of non-degraded gliadin in breast milk from healthy mothers. Scand. J. Gastroenterol. 1998, 33, 1186–1192. [Google Scholar] [CrossRef] [PubMed]
- Benard, A.; Desreumeaux, P.; Huglo, D.; Hoorelbeke, A.; Tonnel, A.B.; Wallaert, B. Increased intestinal permeability in bronchial asthma. J. Allergy Clin. Immunol. 1996, 97, 1173–1178. [Google Scholar] [CrossRef]
- Caffarelli, C.; Cavagni, G.; Menzies, I.S.; Bertolini, P.; Atherton, D.J. Elimination diet and intestinal permeability in atopic eczema: A preliminary study. Clin. Exp. Allergy 1993, 23, 28–31. [Google Scholar] [CrossRef] [PubMed]
- Majamaa, H.; Isolauri, E. Evaluation of the gut mucosal barrier: Evidence for increased antigen transfer in children with atopic eczema. J. Allergy Clin. Immunol. 1996, 97, 985–990. [Google Scholar] [CrossRef]
- Brunekreef, B.; Smit, J.; de Jongste, J.; Neijens, H.; Gerritsen, J.; Postma, D.; Aalberse, R.; Koopman, L.; Kerkhof, M.; Wijga, A.; et al. The Prevention and Incidence of Asthma and Mite Allergy (PIAMA) birth cohort study: Design and first results. Pediatr. Allergy Immunol. 2003, 13, 55–60. [Google Scholar] [CrossRef]
- Wijga, A.H.; Kerkhof, M.; Gehring, U.; de Jongste, J.C.; Postma, D.S.; Aalberse, R.C.; Wolse, A.P.; Koppelman, G.H.; van Rossem, L.; Oldenwening, M.; et al. Cohort profile: The prevention and incidence of asthma and mite allergy (PIAMA) birth cohort. Int. J. Epidemiol. 2014, 43, 527–535. [Google Scholar] [CrossRef] [Green Version]
- Hettinga, K.A.; Reina, F.M.; Boeren, S.; Zhang, L.; Koppelman, G.H.; Postma, D.S.; Vervoort, J.J.; Wijga, A.H. Difference in the breast milk proteome between allergic and nonallergic mothers. PLoS ONE 2015, 10, e0122234. [Google Scholar] [CrossRef] [Green Version]
- Cox, J.; Mann, M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008, 26, 1367–1372. [Google Scholar] [CrossRef]
- Verheggen, K.; Ræder, H.; Berven, F.S.; Martens, L.; Barsnes, H.; Vaudel, M. Anatomy and evolution of database search engines-a central component of mass spectrometry based proteomic workflows. Mass Spectrom. Rev. 2017. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- D’Alessandro, A.; Zolla, L.; Scaloni, A. The bovine milk proteome: Cherishing, nourishing and fostering molecular complexity. An interactomics and functional overview. Mol. Biosyst. 2011, 7, 579–597. [Google Scholar] [CrossRef] [PubMed]
- D’Alessandro, A.; Scaloni, A.; Zolla, L. Human milk proteins: An interactomics and updated functional overview. J Proteome Res 2010, 9, 3339–3373. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Li, Z.; Huang, H.; Suzek, B.E.; Wu, C.H.; UniProt, C. A fast peptide match service for UniProt knowledgebase. Bioinformatics 2013, 29, 2808–2809. [Google Scholar] [CrossRef] [Green Version]
- R Development Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2018. [Google Scholar]
- Høst, A.; Husby, S.; Hansen, L.G.; Osterballe, O. Bovine β-1actoglobulin in human milk from atopic and non-atopic mothers. Relationship to maternal intake of homogenized and unhomogenized milk. Clin. Exp. Allergy 1990, 20, 383–387. [Google Scholar] [CrossRef]
- Axelsson, I.; Jakobsson, I.; Lindberg, T.; Benediktsson, B. Bovine beta-lactoglobulin in the human milk. A longitudinal study during the whole lactation period. Acta Paediatr. Scand. 1986, 75, 702–707. [Google Scholar] [CrossRef]
- Sanchón, J.; Fernández-Tomé, S.; Miralles, B.; Hernández-Ledesma, B.; Tomé, D.; Gaudichon, C.; Recio, I. Protein degradation and peptide release from milk proteins in human jejunum. Comparison with in vitro gastrointestinal simulation. Food Chem. 2018, 239, 486–494. [Google Scholar] [CrossRef]
- European Food Safety Authority. Scientific opinion on dietary reference values for protein. EFSA J. 2012, 10, 2557. [Google Scholar] [CrossRef]
- Liccardi, G.; Asero, R.; D’Amato, M.; D’Amato, G. Role of sensitization to mammalian serum albumin in allergic disease. Curr. Allergy Asthma Rep. 2011, 11, 421–426. [Google Scholar] [CrossRef]
- Zahradnik, E.; Raulf, M. Animal allergens and their presence in the environment. Front. Immunol. 2014, 5, 76. [Google Scholar] [CrossRef] [Green Version]
- Matangkasombut, P.; Padungpak, S.; Thaloengsok, S.; Kamchaisatian, W.; Sasisakulporn, C.; Jotikasthira, W.; Benjaponpitak, S.; Manuyakorn, W. Detection of β-lactoglobulin in human breast-milk 7 days after cow milk ingestion. Paediatr. Int. Child Health 2017, 37, 199–203. [Google Scholar] [CrossRef]
- Sorva, R.; Mäkinen-Kiljunen, S.; Juntunen-Backman, K. β-Lactoglobulin secretion in human milk varies widely after cow’s milk ingestion in mothers of infants with cow’s milk allergy. J. Allergy Clin. Immunol. 1994, 93, 787–792. [Google Scholar] [CrossRef]
- Bertino, E.; Farinasso, D.; Cavaletto, M.; Coscia, A.; Prandi, G.; Giuffrida, M.G.; Costa, S.; Fabris, C.; Conti, A. Absence in human milk of bovine β-lactoglobulin ingested by the mother. Unreliability of ELISA measurements. Acta Biomed. l’Ateneo Parm. 1997, 68, 15–19. [Google Scholar]
- Restani, P.; Gaiaschi, A.; Plebani, A.; Beretta, B.; Velonà, T.; Cavagni, G.; Poiesi, C.; Ugazio, A.G.; Galli, C.L. Evaluation of the presence of bovine proteins in human milk as a possible cause of allergic symptoms in breast-fed children. Ann. Allergy Asthma Immunol. 2000, 84, 353–360. [Google Scholar] [CrossRef]
- Bevilacqua, C.; Montagnac, G.; Benmerah, A.; Candalh, C.; Brousse, N.; Cerf-Bensussan, N.; Perdue, M.H.; Heyman, M. Food allergens are protected from degradation during CD23-mediated transepithelial transport. Int. Arch. Allergy Immunol. 2004, 135, 108–116. [Google Scholar] [CrossRef]
- Caillard, I.; Tomé, D. Transport of β-lactoglobulin and α-lactalbumin in enterocyte-like Caco-2 cells. Reprod. Nutr. Dev. 1995, 35, 179–188. [Google Scholar] [CrossRef] [Green Version]
- Sakurai, N.; Nishio, S.; Akiyama, Y.; Miyata, S.; Oshima, K.; Nadano, D.; Matsuda, T. Apical-to-basolateral transepithelial transport of cow’s milk caseins by intestinal Caco-2 cell monolayers: MS-based quantitation of cellularly degraded a- and b-casein fragments. J. Biochem. 2018, 164, 113–125. [Google Scholar] [CrossRef]
- Rytkönen, J.; Valkonen, K.H.; Virtanen, V.; Foxwell, R.A.; Kyd, J.M.; Cripps, A.W.; Karttunen, T.J. Enterocyte and M-cell transport of native and heat-denatured bovine β-lactoglobulin: Significance of heat denaturation. J. Agric. Food Chem. 2006, 54, 1500–1507. [Google Scholar] [CrossRef]
- Calderón, M.A.; Linneberg, A.; Kleine-Tebbe, J.; De Blay, F.; Hernandez Fernandez De Rojas, D.; Virchow, J.C.; Demoly, P. Respiratory allergy caused by house dust mites: What do we really know? J. Allergy Clin. Immunol. 2015, 136, 38–48. [Google Scholar] [CrossRef] [Green Version]
- Gill, N.; Wlodarska, M.; Finlay, B.B. The future of mucosal immunology: Studying an integrated system-wide organ. Nat. Immunol. 2010, 11, 558–560. [Google Scholar] [CrossRef]
- Zhu, T.H.; Zhu, T.R.; Tran, K.A.; Sivamani, R.K.; Shi, V.Y. Epithelial barrier dysfunctions in atopic dermatitis: A skin–gut–lung model linking microbiome alteration and immune dysregulation. Br. J. Dermatol. 2018, 179, 570–581. [Google Scholar] [CrossRef] [PubMed]
- Hijazi, Z.; Molla, A.M.; Al-Habashi, H.; Muawad, W.M.; Mollo, A.M.; Sharma, P.N. Intestinal permeability is increased in bronchial asthma. Arch. Dis. Child. 2004, 89, 227–229. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tulic, M.K.; Vivinus-Nébot, M.; Rekima, A.; Medeiros, S.R.; Bonnart, C.; Shi, H.; Walker, A.; Dainese, R.; Boyer, J.; Vergnolle, N.; et al. Presence of commensal house dust mite allergen in human gastrointestinal tract: A potential contributor to intestinal barrier dysfunction. Gut 2016, 65, 757–766. [Google Scholar] [CrossRef] [PubMed]
- Ménard, S.; Cerf-Bensussan, N.; Heyman, M. Multiple facets of intestinal permeability and epithelial handling of dietary antigens. Mucosal Immunol. 2010, 3, 247–259. [Google Scholar] [CrossRef]
- Sopo, S.M.; Monaco, S.; Greco, M.; Scala, G. Chronic food protein-induced enterocolitis syndrome caused by cow’s milk proteins passed through breast milk. Int. Arch. Allergy Immunol. 2014, 164, 207–209. [Google Scholar] [CrossRef]
- Vergara Perez, I.; Vila Sexto, L. Suspected severe acute food protein–induced enterocolitis syndrome caused by cow’s milk through breast milk. Ann. Allergy Asthma Immunol. 2018, 121, 245–246. [Google Scholar] [CrossRef]
- Verhasselt, V.; Milcent, V.; Cazareth, J.; Kanda, A.; Fleury, S.; Dombrowicz, D.; Glaichenhaus, N.; Julia, V. Breast milk-mediated transfer of an antigen induces tolerance and protection from allergic asthma. Nat. Med. 2008, 14, 170–175. [Google Scholar] [CrossRef]
- Gouw, J.W.; Jo, J.; Meulenbroek, L.A.P.M.; Heijjer, T.S.; Kremer, E.; Sandalova, E.; Knulst, A.C.; Jeurink, P.V.; Garssen, J.; Rijnierse, A.; et al. Identification of peptides with tolerogenic potential in a hydrolysed whey-based infant formula. Clin. Exp. Allergy 2018, 48, 1345–1353. [Google Scholar] [CrossRef] [Green Version]
- Peters, R.L.; Koplin, J.J.; Dharmage, S.C.; Tang, M.L.; McWilliam, V.L.; Gurrin, L.C.; Neeland, M.R.; Lowe, A.J.; Ponsonby, A.L.; Allen, K.J. Early exposure to cow’s milk protein is associated with a reduced risk of cow’s milk allergic outcomes. J. Allergy Clin. Immunol. Pract. 2019, 7, 462–470. [Google Scholar] [CrossRef]
- Warshaw, A.L.; Walker, W.A.; Isselbacher, K.J. Protein uptake by the intestine: Evidence for absorption of intact macromolecules. Gastroenterology 1974, 66, 987–992. [Google Scholar] [CrossRef]



| Characteristic | Type | Nonallergic | Allergic |
|---|---|---|---|
| House dust mite allergy | Self report | 0 | 7 |
| Doctor diagnosed | 0 | 7 | |
| House dust allergy | Self report | 0 | 8 |
| Doctor diagnosed | 0 | 6 | |
| Allergic to pets | Self report | 0 | 9 |
| Doctor diagnosed | 0 | 8 | |
| Asthma | Self report | 0 | 7 |
| Doctor diagnosed | 0 | 7 | |
| House dust mite Der p IgE (Rast-class) | Class 3 | NA a | 4 |
| Class 4 | NA a | 5 | |
| Class 5 | NA a | 1 | |
| Rhinitis/hay fever | Self report | 0 | 9 |
| Cat as pet in the household | Presence | 3 | 3 |
| Consumption of milk during lactation | Not at all | 2 | 3 |
| 1–3 × a month | 0 | 1 | |
| 1 × a week | 0 | 0 | |
| 2–4 × a week | 0 | 3 | |
| More than 4 × a week | 0 | 0 | |
| 1 × a day | 1 | 1 | |
| Multiple times a day | 7 | 2 | |
| Consumption of milk products during lactation | Not at all | 2 | 0 |
| 1–3 × a month | 0 | 0 | |
| 1 × a week | 0 | 0 | |
| 2–4 × a week | 1 | 0 | |
| More than 4 × a week | 0 | 1 | |
| 1 × a day | 4 | 4 | |
| Multiple times a day | 3 | 5 |
| Sequence | Leading Proteins | Protein Names | Allergic | Nonallergic | Digestion |
|---|---|---|---|---|---|
| ALPMHIR a | B5B0D4 | Beta-lactoglobulin | 10 | 2 | trypsin |
| IDALNENK a | B5B0D4 | Beta-lactoglobulin | 10 | 5 | trypsin |
| LIVTQTMK a | B5B0D4 | Beta-lactoglobulin | 10 | 4 | trypsin |
| LSFNPTQLEEQCHI b | B5B0D4 | Beta-lactoglobulin | 10 | 6 | trypsin |
| TKIPAVFK a | B5B0D4 | Beta-lactoglobulin | 10 | 0 | trypsin |
| TPEVDDEALEK a | B5B0D4 | Beta-lactoglobulin | 10 | 2 | trypsin |
| TPEVDDEALEKFDK a | B5B0D4 | Beta-lactoglobulin | 10 | 5 | trypsin |
| VLVLDTDYKK a | B5B0D4 | Beta-lactoglobulin | 10 | 5 | trypsin |
| VYVEELKPTPEGDLEILLQK a | B5B0D4 | Beta-lactoglobulin | 9 | 1 | trypsin |
| WENDECAQK b | B5B0D4 | Beta-lactoglobulin | 9 | 1 | trypsin |
| WENDECAQKK b | B5B0D4 | Beta-lactoglobulin | 4 | 0 | trypsin |
| SLAMAASDISLLDAQSAPLR b | B5B0D4 | Beta-lactoglobulin | 6 | 0 | semi-specific |
| HHIELRWK | E1BFN5 | Uncharacterized protein | 9 | 8 | trypsin |
| QKYGVVKENVIDLTK | E1BJP1, G3MZU3 | Uncharacterized proteins | 0 | 9 | semi-specific |
| EKESLGWQK | E1BKT9 | Desmoplakin | 0 | 2 | unspecific |
| EHLYQENQYLEQENTQ | E1BMB1 | Ninein | 0 | 6 | unspecific |
| QEELENRTSETNTPQGNQEY | E1BMB1 | Ninein | 8 | 3 | unspecific |
| HEQGMDQDKN | F1MV51 | APC, WNT signalling pathway regulator | 10 | 10 | unspecific |
| SSLSDIDQENNNNK | F1MV51 | APC, WNT signalling pathway regulator | 2 | 3 | unspecific |
| TLQIAEIKDNSGPRSNED | F1MV51 | APC, WNT signalling pathway regulator | 0 | 2 | unspecific |
| QNLAFVSMLNDIAAP | F1N647 | Fatty acid synthase | 0 | 1 | unspecific |
| IQQNSSTTEKI | F2FB38 | Mucin-16 | 6 | 9 | unspecific |
| KFNITDTLMQ | F2FB38 | Mucin-16 | 0 | 1 | unspecific |
| LDQWLCEKL b | P00711 | Alpha-lactalbumin | 4 | 0 | trypsin |
| NICNISCDKFLDD | P00711 | Alpha-lactalbumin | 0 | 1 | unspecific |
| EKVNELSK a | P02662 | Alpha-S1-casein | 7 | 1 | trypsin |
| FFVAPFPEVFGK a | P02662 | Alpha-S1-casein | 2 | 3 | trypsin |
| HIQKEDVPSER a | P02662 | Alpha-S1-casein | 10 | 8 | trypsin |
| HQGLPQEVLNENLLR a | P02662 | Alpha-S1-casein | 5 | 8 | trypsin |
| YLGYLEQLLR a | P02662 | Alpha-S1-casein | 2 | 5 | trypsin |
| SCQAQPTTMAR b | P02668 | Kappa-casein | 9 | 3 | trypsin |
| AEFVEVTK a | P02769 | Serum albumin | 7 | 10 | trypsin |
| DAFLGSFLYEYSR a | P02769 | Serum albumin | 6 | 4 | trypsin |
| DLGEEHFK b | P02769 | Serum albumin | 0 | 9 | trypsin |
| DTHKSEIAHR a | P02769 | Serum albumin | 0 | 10 | trypsin |
| DVCKNYQEAK b | P02769 | Serum albumin | 10 | 10 | trypsin |
| FKDLGEEHFK a | P02769 | Serum albumin | 10 | 10 | trypsin |
| HLVDEPQNLIK a | P02769 | Serum albumin | 4 | 9 | trypsin |
| LVNELTEFAK a | P02769 | Serum albumin | 7 | 10 | trypsin |
| QNCDQFEK b | P02769 | Serum albumin | 0 | 5 | trypsin |
| RHPEYAVSVLLR a | P02769 | Serum albumin | 7 | 10 | trypsin |
| SLHTLFGDELCK b | P02769 | Serum albumin | 1 | 8 | trypsin |
| TCVADESHAGCEK b | P02769 | Serum albumin | 2 | 7 | trypsin |
| GKYLYEIAR | P02769 | Serum albumin | 9 | 10 | semi-specific |
| KQTALVELLK b | P02769 | Serum albumin | 2 | 5 | unspecific |
| IKVMNDLSPKSNLR | P07353 | Interferon gamma | 2 | 1 | semi-specific |
| DLKLVEQQNPK | P08037 | Beta-1,4-galactosyltransferase 1 | 0 | 2 | semi-specific |
| AQFVPLPVSVSVEFAVAATDCIAK b | P12763 | Alpha-2-HS-glycoprotein | 9 | 0 | trypsin |
| VNLLVDRQWQAVRNR | P15396 | Ectonucleotide pyrophosphatase | 10 | 10 | trypsin |
| KLLNNITNDLR | P21758 | Macrophage scavenger receptor | 4 | 0 | unspecific |
| NLLFNDNTECLAK b | P24627 | Lactotransferrin | 4 | 1 | trypsin |
| NKHSNLIESQENSK | P31098, P31096 | Osteopontin-K, Osteopontin | 9 | 7 | trypsin |
| NVTRQAYWQIHMDQ | P80209 | Cathepsin D | 0 | 3 | unspecific |
| NGNNPNCCMNQK | P80457 | Xanthine dehydrogenase/oxidase | 1 | 0 | semi-specific |
| EKQLPNGDWPQENISGVFNKSCA | P84466 | Lanosterol synthase | 5 | 3 | unspecific |
| VSITCSGSSSNIGR b | Q1RMN8 | Immunoglobulin light chain | 8 | 5 | trypsin |
| CASFRENVLR b | Q29443 | Serotransferrin | 10 | 10 | trypsin |
| QMERALLENE | Q2HJ49 | Moesin | 0 | 3 | semi-specific |
| NGEGQVLFETEISR | Q2TBX4 | Heat shock 70 kDa protein 13 | 3 | 8 | trypsin |
| NIIKSGSDEVQ | Q2UVX4 | Complement C3 | 1 | 0 | unspecific |
| VALNKLK | Q58D55 | Beta-galactosidase | 2 | 0 | trypsin |
| VYVEQLKPTPEGDLEILLQK | Q9BDG3 | Beta lactoglobulin D | 1 | 0 | trypsin |
| Sequence | Leading Proteins | Leading Organisms or LCA b | Allergic | Nonallergic | Digestion |
|---|---|---|---|---|---|
| QNWASLQPYKKL | Q08169, A0A0M9A8V0, I1VC83, A0A2A3EHG0, Q95PD7, A0A0L7RCK4, A0A310SIY9 | Apidae (family) (bees) | 1 | 2 | semi-specific |
| RPSHQQPR | P43237, N1NEW2 | Arachis (genus) (legumes) | 6 | 4 | trypsin |
| MQDQLDQVQK | Q8MUF6, Q9BMM8, A0A1B2YLJ8 | Astigmatina (cohort) (mites) | 1 | 5 | unspecific |
| KELKKKVEADGEND | A0A2V1CGL9 | Cadophora sp. DSE1049 | 6 | 4 | unspecific |
| QIANSDEVEKI | Q24702 | Dictyocaulus viviparus | 3 | 6 | unspecific |
| KCAADESAENCDK | P35747 | Equus caballus | 7 | 3 | trypsin |
| LVNEVTEFAKK a | P35747 | Equus caballus | 10 | 8 | trypsin |
| KEPERNECFLQHK a | P49064 | Felis catus | 5 | 8 | trypsin |
| PCFSALQVDETYVPK | P49064 | Felis catus | 1 | 0 | trypsin |
| YICENQDSISTK | P49064 | Felis catus | 0 | 5 | trypsin |
| SALQVDETYVPK | P49064 | Felis catus | 3 | 4 | semi-specific |
| KEQVARFTAGTNPK | A9QQ26 | Lycosa singoriensis | 10 | 10 | trypsin |
| EQVQELR | A0A1L8GUE3, A0A3Q0GE46, A0A151P804 | Tetrapoda (superclass) (4-limbed vertebrates) | 2 | 2 | trypsin |
| QQQTLQQILQQQ | P04723 | Triticum aestivum | 10 | 10 | unspecific |
| QVLQQSSYQQLQQ | P04723 | Triticum aestivum | 0 | 2 | unspecific |
| QFKPEEMTNIIK | P35083, A4KA55 | Zea mays | 8 | 4 | semi-specific |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dekker, P.M.; Boeren, S.; Wijga, A.H.; Koppelman, G.H.; Vervoort, J.J.M.; Hettinga, K.A. Maternal Allergy and the Presence of Nonhuman Proteinaceous Molecules in Human Milk. Nutrients 2020, 12, 1169. https://0-doi-org.brum.beds.ac.uk/10.3390/nu12041169
Dekker PM, Boeren S, Wijga AH, Koppelman GH, Vervoort JJM, Hettinga KA. Maternal Allergy and the Presence of Nonhuman Proteinaceous Molecules in Human Milk. Nutrients. 2020; 12(4):1169. https://0-doi-org.brum.beds.ac.uk/10.3390/nu12041169
Chicago/Turabian StyleDekker, Pieter M., Sjef Boeren, Alet H. Wijga, Gerard H. Koppelman, Jacques J. M. Vervoort, and Kasper A. Hettinga. 2020. "Maternal Allergy and the Presence of Nonhuman Proteinaceous Molecules in Human Milk" Nutrients 12, no. 4: 1169. https://0-doi-org.brum.beds.ac.uk/10.3390/nu12041169