Unique Genetic and Histological Signatures of Mouse Pericardial Adipose Tissue
Abstract
:1. Introduction
2. Methods
2.1. Animal and Experimental Design
2.2. RNA Isolation
2.3. cDNA Synthesis and qRT-PCR
2.4. Histology
2.5. Statistical Analysis
3. Results
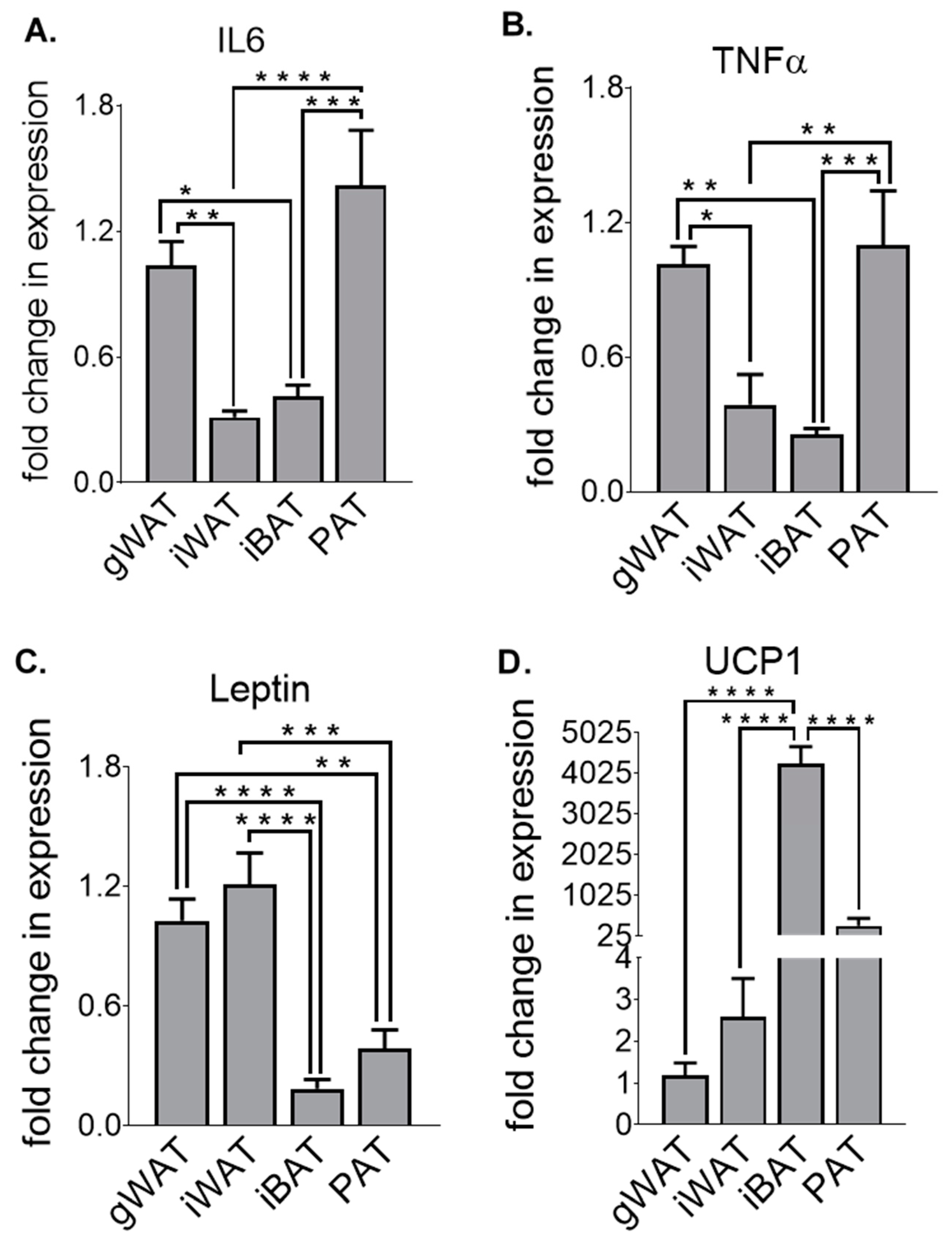
3.1. Adipose Tissue Gene Expression Differences at Baseline in Chow-Fed Mice
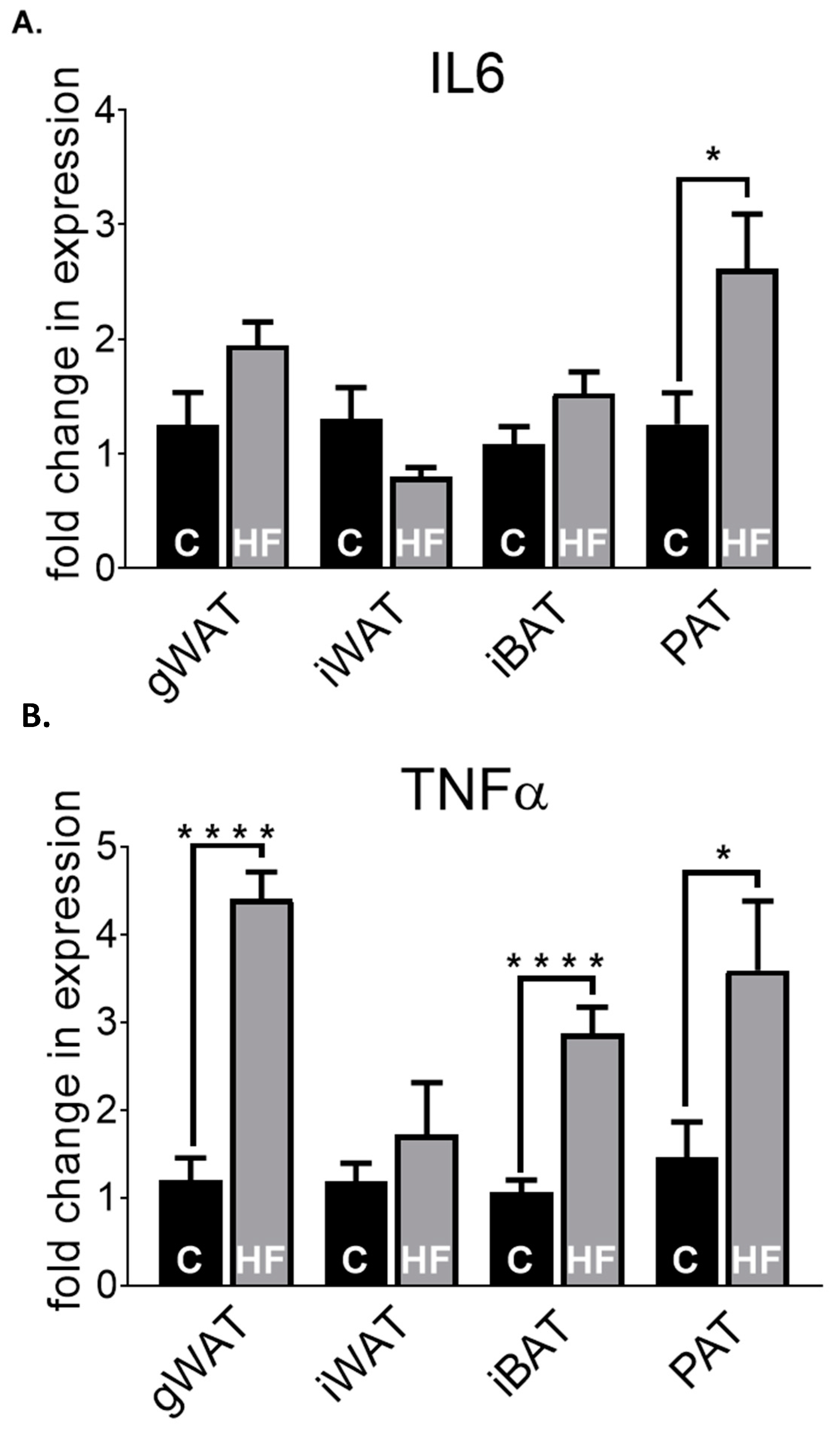
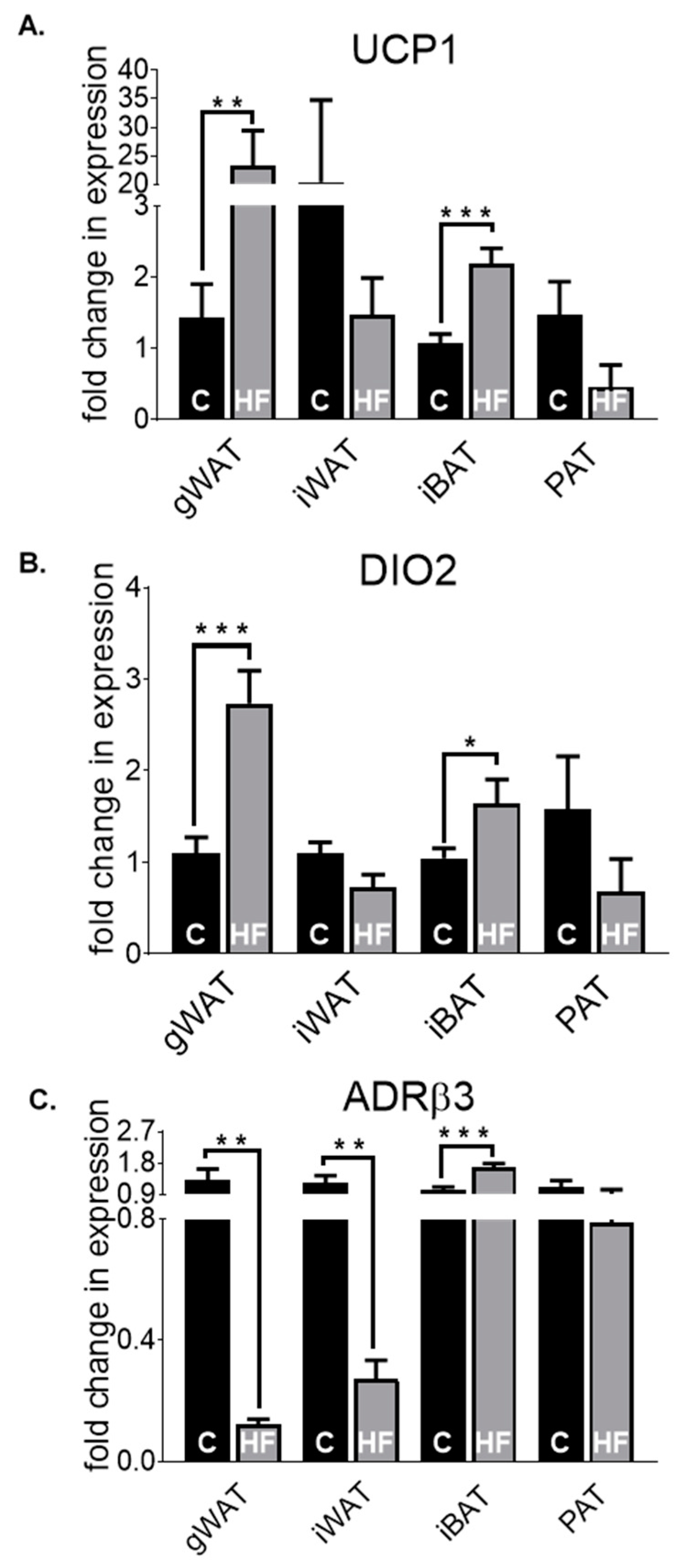
3.2. Adipose Tissue Gene Expression Differences in Chow-Fed Mice and HF-Fed Mice
3.3. Adipose Tissue Gene Expression Differences in HF-Fed Mice
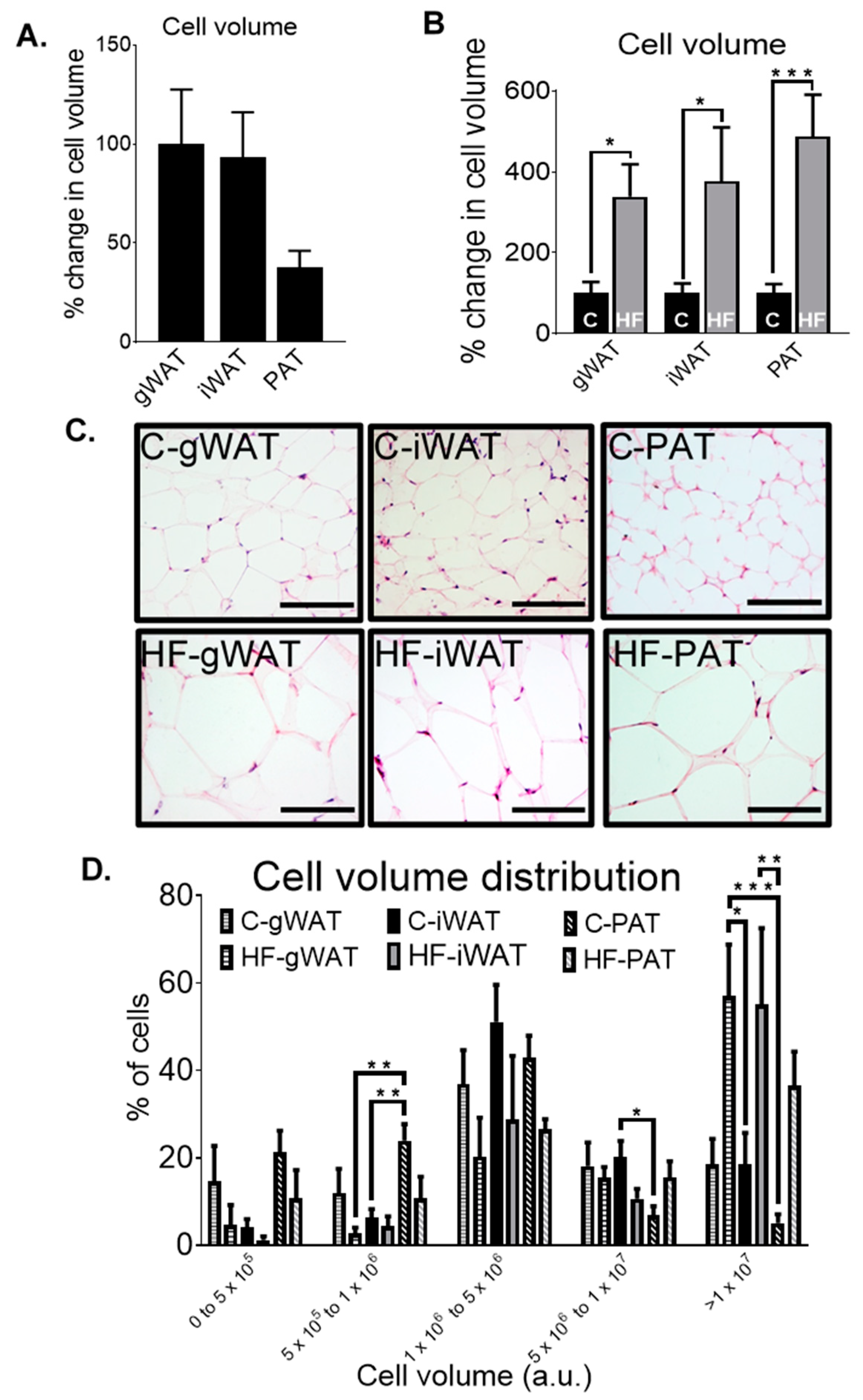
3.4. Adipose Tissue Histological Differences in Chow-Fed and HF-Fed Mice
3.5. The Relationship between Biochemical and Metabolic Parameters, Gene Expression Levels and Cell Volume in Adipose Tissue in Chow- and HF-Fed Mice
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Cornier, M.; Després, J.; Davis, N.; Grossniklaus, D.A.; Klein, S.; Lamarche, B.; Lopez-Jimenez, F.; Rao, G.; St-Onge, M.; Towfighi, A.; et al. Assessing adiposity: A scientific statement from the American Heart Association. Circulation 2011, 124, 1996–2019. [Google Scholar] [CrossRef] [Green Version]
- Montani, J.-P.; Carroll, J.F.; Dwyer, T.M.; Antic, V.; Yang, Z.; Dulloo, A.G. Ectopic fat storage in heart, blood vessels and kidneys in the pathogenesis of cardiovascular diseases. Int. J. Obes. 2004, 28 (Suppl. 4), S58–S65. [Google Scholar] [CrossRef] [Green Version]
- Gustafson, B.; Smith, U. Regulation of white adipogenesis and its relation to ectopic fat accumulation and cardiovascular risk. Atherosclerosis 2015, 241, 27–35. [Google Scholar] [CrossRef] [PubMed]
- Chu, A.Y.; Deng, X.; A Fisher, V.; Drong, A.; Zhang, Y.; Feitosa, M.F.; Liu, C.-T.; Weeks, O.; Choh, A.C.; Duan, Q.; et al. Multiethnic genome-wide meta-analysis of ectopic fat depots identifies loci associated with adipocyte development and differentiation. Nat. Genet. 2016, 49, 125–130. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ravussin, E.; Galgani, J.E. The implication of brown adipose tissue for humans. Annu. Rev. Nutr. 2011, 31, 33–47. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bjorndal, B.; Burri, L.; Staalesen, V.; Skorve, J.; Berge, R.K. Different Adipose Depots: Their Role in the Development of Metabolic Syndrome and Mitochondrial Response to Hypolipidemic Agents. J. Obes. 2011, 2011, 490650. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guglielmi, V.; Sbraccia, P. Obesity phenotypes: Depot-differences in adipose tissueand their clinical implications. Eat. Weight Disord. 2018, 23, 3–14. [Google Scholar] [CrossRef]
- Ibrahim, M.M. Subcutaneous and visceral adipose tissue: Structural and functional differences. Obes. Rev. 2010, 11, 11–18. [Google Scholar] [CrossRef]
- Wajchenberg, B.L.; Giannella-Neto, D.; Da Silva, M.E.; Santos, R.F. Depot-Specific Hormonal Characteristics of Subcutaneous and Visceral Adipose Tissue and their Relation to the Metabolic Syndrome. Horm. Metab. Res. 2002, 34, 616–621. [Google Scholar] [CrossRef]
- Iacobellis, G. Epicardial and pericardial fat: Close, but very different. Obesity 2009, 17, 626–627. [Google Scholar] [CrossRef]
- Kaushik, M.; Reddy, Y.M. Distinction of “fat around the heart”. J. Am. Coll. Cardiol. 2011, 58, 1640–1641. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iozzo, P. Myocardial, Perivascular, and Epicardial Fat. Diabetes Care 2011, 34 (Suppl. 2), S371–S379. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Britton, K.A.; Fox, C.S. Ectopic Fat Depots and Cardiovascular Disease. Circulation 2011, 124, e837–e841. [Google Scholar] [CrossRef] [PubMed]
- Iacobellis, G. Local and systemic effects of the multifaceted epicardial adipose tissue depot. Nat. Rev. Endocrinol. 2015, 11, 363–371. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Zhang, A.; Hamilton, D.J.; Deng, T. Epicardial Fat in the Maintenance of Cardiovascular Health. Methodist DeBakey Cardiovasc. J. 2017, 13, 20–24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salazar, J.; Luzardo, E.; Mejías, J.C.; Rojas, M.; Ferreira, A.; Rivas-Ríos, J.R.; Bermúdez, V. Epicardial Fat: Physiological, Pathological, and Therapeutic Implications. Cardiol. Res. Pract. 2016, 2016, 1291537. [Google Scholar] [CrossRef] [Green Version]
- Aslanabadi, N.; Salehi, R.; Javadrashid, A.; Tarzamani, M.; Khodadad, B.; Enamzadeh, E.; Montazerghaem, H. Epicardial and Pericardial Fat Volume Correlate with the Severity of Coronary Artery Stenosis. J. Cardiovasc. Thorac. Res. 2014, 6, 235–239. [Google Scholar] [CrossRef] [Green Version]
- Mohammadzadeh, M.; Mohammadzadeh, V.; Shakiba, M.; Motevalli, M.; Abedini, A.; Kadivar, S.; Entezari, P.; Mohammadzadeh, A. Assessing the Relation of Epicardial Fat Thickness and Volume, Quantified by 256-Slice Computed Tomography Scan, With Coronary Artery Disease and Cardiovascular Risk Factors. Arch. Iran. Med. 2018, 21, 95–100. [Google Scholar]
- McClain, J.; Hsu, F.; Brown, E.; Burke, G.; Carr, J.J.; Harris, T.; Kritchevsky, S.; Szklo, M.; Tracy, R.; Ding, J.; et al. Pericardial adipose tissue and coronary artery calcification in the Multi-ethnic Study of Atherosclerosis (MESA). Obesity 2013, 21, 1056–1063. [Google Scholar] [CrossRef] [Green Version]
- Ding, J.; Hsu, F.-C.; Harris, T.B.; Liu, Y.; Kritchevsky, S.; Szklo, M.; Ouyang, P.; A Espeland, M.; Lohman, K.K.; Criqui, M.H.; et al. The association of pericardial fat with incident coronary heart disease: The Multi-Ethnic Study of Atherosclerosis (MESA). Am. J. Clin. Nutr. 2009, 90, 499–504. [Google Scholar] [CrossRef]
- Chechi, K.; Vijay, J.; Voisine, P.; Mathieu, P.; Bossé, Y.; Tchernof, A.; Grundberg, E.; Richard, D. UCP1 expression-associated gene signatures of human epicardial adipose tissue. JCI Insight 2019, 4. [Google Scholar] [CrossRef] [PubMed]
- Fox, C.S.; White, C.C.; Lohman, K.; Heard-Costa, N.; Cohen, P.; Zhang, Y.; Johnson, A.D.; Emilsson, V.; Liu, C.-T.; Chen, Y.-D.I.; et al. Genome-Wide Association of Pericardial Fat Identifies a Unique Locus for Ectopic Fat. PLoS Genet. 2012, 8, e1002705. [Google Scholar] [CrossRef]
- Bale, L.K.; West, S.A.; Conover, C.A. Characterization of mouse pericardial fat: Regulation by PAPP-A. Growth Horm. IGF Res. 2018, 42, 1–7. [Google Scholar] [CrossRef]
- Fox, C.S.; Gona, P.; Hoffmann, U.; Porter, S.A.; Salton, C.J.; Massaro, J.M.; Levy, D.; Larson, M.G.; D’Agostino, R.B.; O’Donnell, C.J.; et al. Pericardial fat, intrathoracic fat, and measures of left ventricular structure and function: The Framingham Heart Study. Circulation 2009, 119, 1586–1591. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cannon, B.; Nedergaard, J. Brown Adipose Tissue: Function and Physiological Significance. Physiol. Rev. 2004, 84, 277–359. [Google Scholar] [CrossRef] [PubMed]
- Diaz, M.B.; Herzig, S.; Vegiopoulos, A. Thermogenic adipocytes: From cells to physiology and medicine. Metabolism 2014, 63, 1238–1249. [Google Scholar] [CrossRef] [PubMed]
- Virtanen, K.A.; Lidell, M.E.; Orava, J.; Heglind, M.; Westergren, R.; Niemi, T.; Taittonen, M.; Laine, J.; Savisto, N.-J.; Enerbäck, S.; et al. Functional Brown Adipose Tissue in Healthy Adults. N. Engl. J. Med. 2009, 360, 1518–1525. [Google Scholar] [CrossRef] [PubMed]
- Cypess, A.M.; Lehman, S.; Williams, G.; Tal, I.; Rodman, D.; Goldfine, A.B.; Kuo, F.C.; Palmer, E.L.; Tseng, Y.-H.; Doria, A.; et al. Identification and importance of brown adipose tissue in adult humans. N. Engl. J. Med. 2009, 360, 1509–1517. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nedergaard, J.; Bengtsson, T.; Cannon, B. Unexpected evidence for active brown adipose tissue in adult humans. Am. J. Physiol. Metab. 2007, 293, E444–E452. [Google Scholar] [CrossRef] [PubMed]
- Alcalá, M.; Calderon-Dominguez, M.; Serra, D.; Herrero, L.; Viana, M. Mechanisms of Impaired Brown Adipose Tissue Recruitment in Obesity. Front. Physiol. 2019, 10, 94. [Google Scholar] [CrossRef]
- Shabalina, I.; Petrović, N.; De Jong, J.M.A.; Kalinovich, A.V.; Cannon, B.; Nedergaard, J. UCP1 in Brite/Beige Adipose Tissue Mitochondria Is Functionally Thermogenic. Cell Rep. 2013, 5, 1196–1203. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Waldén, T.B.; Hansen, I.R.; Timmons, J.; Cannon, B.; Nedergaard, J. Recruited vs. nonrecruited molecular signatures of brown, “brite,” and white adipose tissues. Am. J. Physiol. Metab. 2012, 302, E19–E31. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Christoffolete, M.A.; Linardi, C.C.; De Jesus, L.; Ebina, K.N.; Carvalho, S.D.; Ribeiro, M.O.; Rabelo, R.; Curcio, C.; Martins, L.; Kimura, E.T.; et al. Mice with targeted disruption of the Dio2 gene have cold-induced overexpression of the uncoupling protein 1 gene but fail to increase brown adipose tissue lipogenesis and adaptive thermogenesis. Diabetes 2004, 53, 577–584. [Google Scholar] [CrossRef] [Green Version]
- Westerterp, K.R. Diet induced thermogenesis. Nutr. Metab. 2004, 1, 5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hibi, M.; Oishi, S.; Matsushita, M.; Yoneshiro, T.; Yamaguchi, T.; Usui, C.; Yasunaga, K.; Katsuragi, Y.; Kubota, K.; Tanaka, S.; et al. Brown adipose tissue is involved in diet-induced thermogenesis and whole-body fat utilization in healthy humans. Int. J. Obes. 2016, 40, 1655–1661. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lowell, B.B.; Wada, T.; Kuzuyama, T.; Satoh, S.; Kuramitsu, S.; Yokoyama, S.; Unzai, S.; Tame, J.R.H.; Park, S.-Y.; Bachman, E.S. β-Adrenergic Receptors, Diet-induced Thermogenesis, and Obesity. J. Boil. Chem. 2003, 278, 29385–29388. [Google Scholar] [CrossRef] [Green Version]
- Feldmann, H.M.; Golozoubova, V.; Cannon, B.; Nedergaard, J. UCP1 Ablation Induces Obesity and Abolishes Diet-Induced Thermogenesis in Mice Exempt from Thermal Stress by Living at Thermoneutrality. Cell Metab. 2009, 9, 203–209. [Google Scholar] [CrossRef] [Green Version]
- Bradley, D.; Liu, J.; Blaszczak, A.; Wright, V.; Jalilvand, A.; Needleman, B.; Noria, S.; Renton, D.; Hsueh, W. Adipocyte DIO2 Expression Increases in Human Obesity but Is Not Related to Systemic Insulin Sensitivity. J. Diabetes Res. 2018, 2018, 2464652. [Google Scholar] [CrossRef]
- Elie, A.G.I.M.; Jensen, P.S.; Nissen, K.D.; Geraets, I.M.E.; Xu, A.; Song, E.; Hansen, M.L.; Irmukhamedov, A.; Rasmussen, L.M.; Wang, Y.; et al. Adipokine Imbalance in the Pericardial Cavity of Cardiac and Vascular Disease Patients. PLoS ONE 2016, 11, e0154693. [Google Scholar] [CrossRef] [Green Version]
- Berg, A.H.; Scherer, P.E. Adipose Tissue, Inflammation, and Cardiovascular Disease. Circ. Res. 2005, 96, 939–949. [Google Scholar] [CrossRef] [Green Version]
- Cinti, S.; Frederich, R.C.; Zingaretti, M.C.; Matteis, R.D.L.; Flier, J.S.; Lowell, B.B. Immunohistochemical localization of leptin and uncoupling protein in white and brown adipose tissue. Endocrinology 1997, 138, 797–804. [Google Scholar] [CrossRef] [PubMed]
- Flier, J.S. Leptin expression and action: New experimental paradigms. Proc. Natl. Acad. Sci. USA 1997, 94, 4242–4245. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, D.W.; Kim, B.S.; Kwon, H.-S.; Gil Kim, C.; Lee, H.-W.; Choi, W.H.; Kim, C.G. Atrophy of brown adipocytes in the adult mouse causes transformation into white adipocyte-like cells. Exp. Mol. Med. 2003, 35, 518–526. [Google Scholar] [CrossRef] [Green Version]
- Cancello, R.; Zingaretti, M.C.; Sarzani, R.; Ricquier, D.; Cinti, S. Leptin and UCP1 genes are reciprocally regulated in brown adipose tissue. Endocrinology 1998, 139, 4747–4750. [Google Scholar] [CrossRef] [PubMed]
- Drolet, R.; Richard, C.; Sniderman, A.D.; Mailloux, J.; Fortier, M.; Huot, C.; Rhéaume, C.; Tchernof, A.; Rh, C. Hypertrophy and hyperplasia of abdominal adipose tissues in women. Int. J. Obes. 2007, 32, 283–291. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Horckmans, M.; Bianchini, M.; Santovito, D.; Megens, R.T.; Springael, J.-Y.; Negri, I.; Vacca, M.; Di Eusanio, M.; Moschetta, A.; Weber, C.; et al. Pericardial Adipose Tissue Regulates Granulopoiesis, Fibrosis, and Cardiac Function After Myocardial Infarction. Circulation 2018, 137, 948–960. [Google Scholar] [CrossRef]
- Kang, Y.M.; Jung, C.H.; Cho, Y.K.; Jang, J.E.; Hwang, J.Y.; Kim, E.H.; Lee, W.J.; Park, J.-Y.; Kim, H.-K. Visceral adiposity index predicts the conversion of metabolically healthy obesity to an unhealthy phenotype. PLoS ONE 2017, 12, e0179635. [Google Scholar] [CrossRef]
- Gabriely, I.; Ma, X.H.; Yang, X.M.; Atzmon, G.; Rajala, M.W.; Berg, A.H.; Scherer, P.; Rossetti, L.; Barzilai, N. Removal of visceral fat prevents insulin resistance and glucose intolerance of aging: An adipokine-mediated process? Diabetes 2002, 51, 2951–2958. [Google Scholar] [CrossRef] [Green Version]



| Gene | Assay ID |
|---|---|
| AdipoQ | Mm00456425_m1 |
| ADRβ3 | Mm02601819_g1 |
| C/EBPα | Mm00514283_s1 |
| COX7A1 | Mm00438297_g1 |
| COX8B | Mm00432648_m1 |
| DIO2 | Mm00515664_m1 |
| FABP4 | Mm00445878_m1 |
| IL6 | Mm00446190_m1 |
| Leptin | Mm00434759_m1 |
| PGC1α | Mm01208835_m1 |
| PPARγ | Mm00440940_m1 |
| PPIA | Mm02342430_g1 |
| TNFα | Mm00443258_m1 |
| UCP1 | Mm01244861_m1 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Al-Dibouni, A.; Gaspar, R.; Ige, S.; Boateng, S.; Cagampang, F.R.; Gibbins, J.; Cox, R.D.; Sellayah, D. Unique Genetic and Histological Signatures of Mouse Pericardial Adipose Tissue. Nutrients 2020, 12, 1855. https://0-doi-org.brum.beds.ac.uk/10.3390/nu12061855
Al-Dibouni A, Gaspar R, Ige S, Boateng S, Cagampang FR, Gibbins J, Cox RD, Sellayah D. Unique Genetic and Histological Signatures of Mouse Pericardial Adipose Tissue. Nutrients. 2020; 12(6):1855. https://0-doi-org.brum.beds.ac.uk/10.3390/nu12061855
Chicago/Turabian StyleAl-Dibouni, A., R. Gaspar, S. Ige, S. Boateng, F. R. Cagampang, J. Gibbins, R. D. Cox, and D. Sellayah. 2020. "Unique Genetic and Histological Signatures of Mouse Pericardial Adipose Tissue" Nutrients 12, no. 6: 1855. https://0-doi-org.brum.beds.ac.uk/10.3390/nu12061855





