FXR, a Key Regulator of Lipid Metabolism, Is Inhibited by ER Stress-Mediated Activation of JNK and p38 MAPK in Large Yellow Croakers (Larimichthys crocea) Fed High Fat Diets
Abstract
:1. Introduction
2. Method
2.1. Animal Feeding Experiment
2.2. Cell Culture and Treatment
2.3. Quantitative Real-Time PCR
2.4. Plasmid Construction and Luciferase Reporter Assay
2.5. Western Blotting
2.6. Chromatin Immunoprecipitation Assay
2.7. Immunoprecipitation
2.8. Statistical Analysis
3. Result
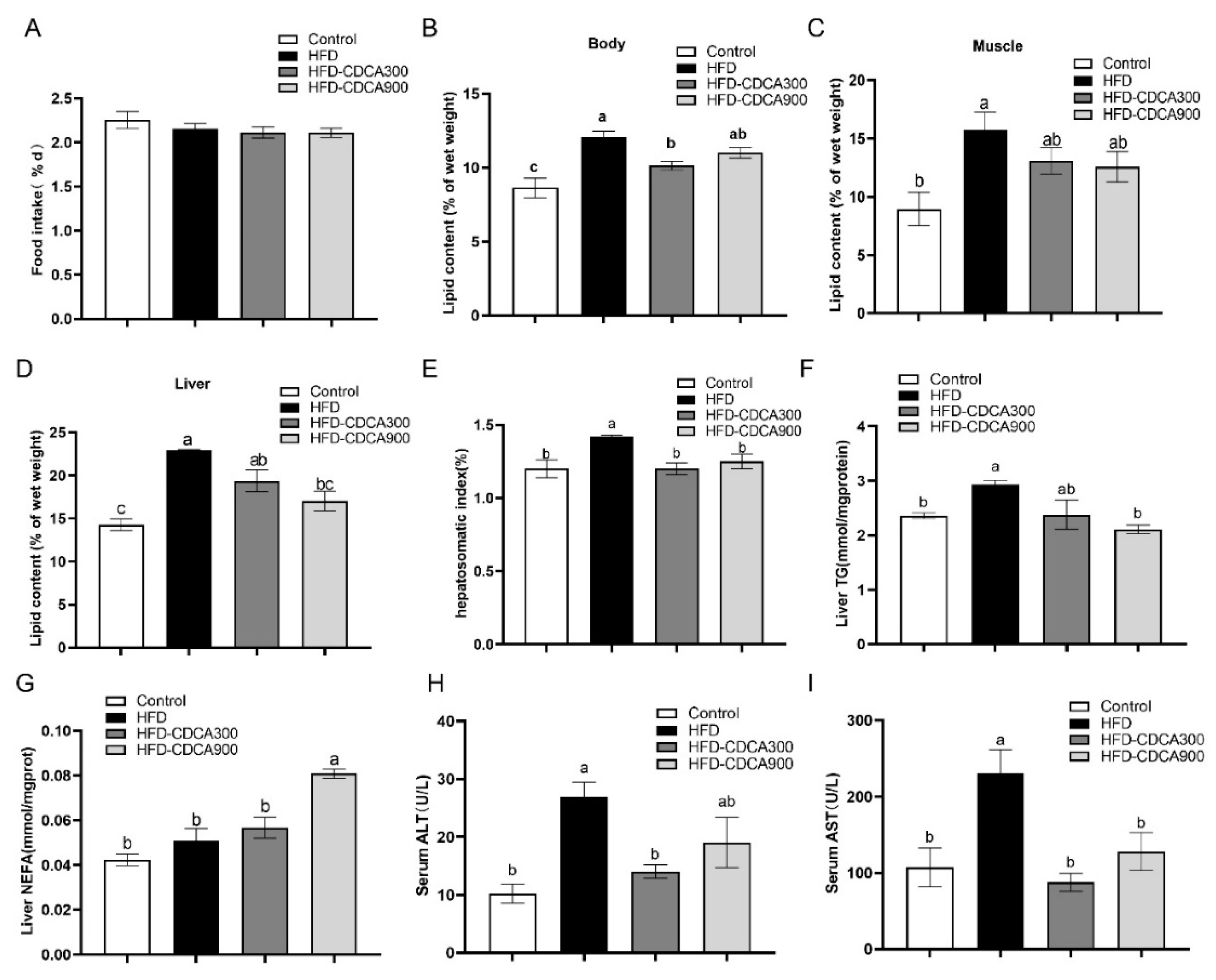
3.1. CDCA Supplementation Decreases HFD-Induced Lipid Deposition in the Liver
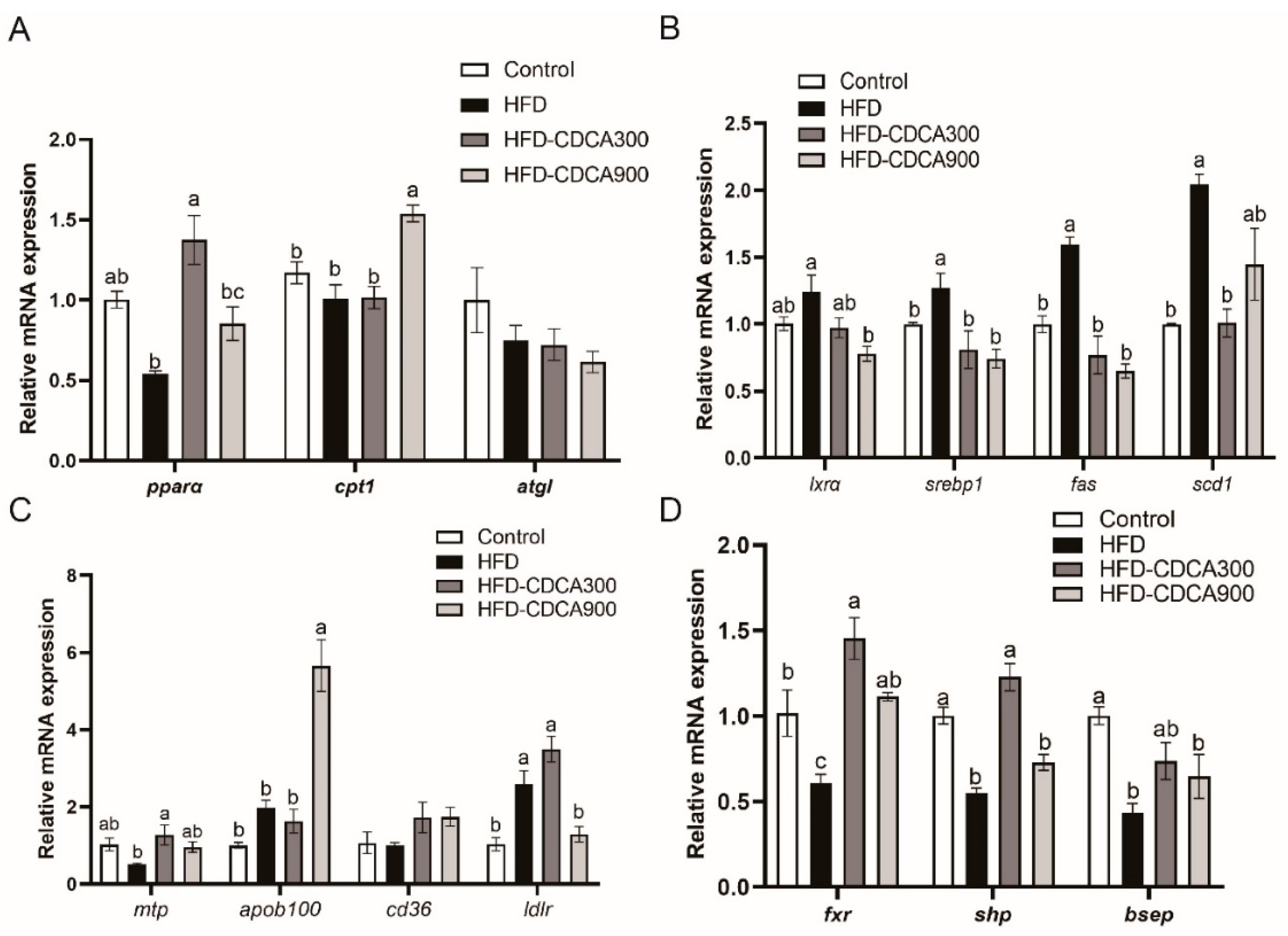
3.2. Effects of HFD and CDCA Supplementation on Lipid Metabolism-Related Gene Expression in the Liver
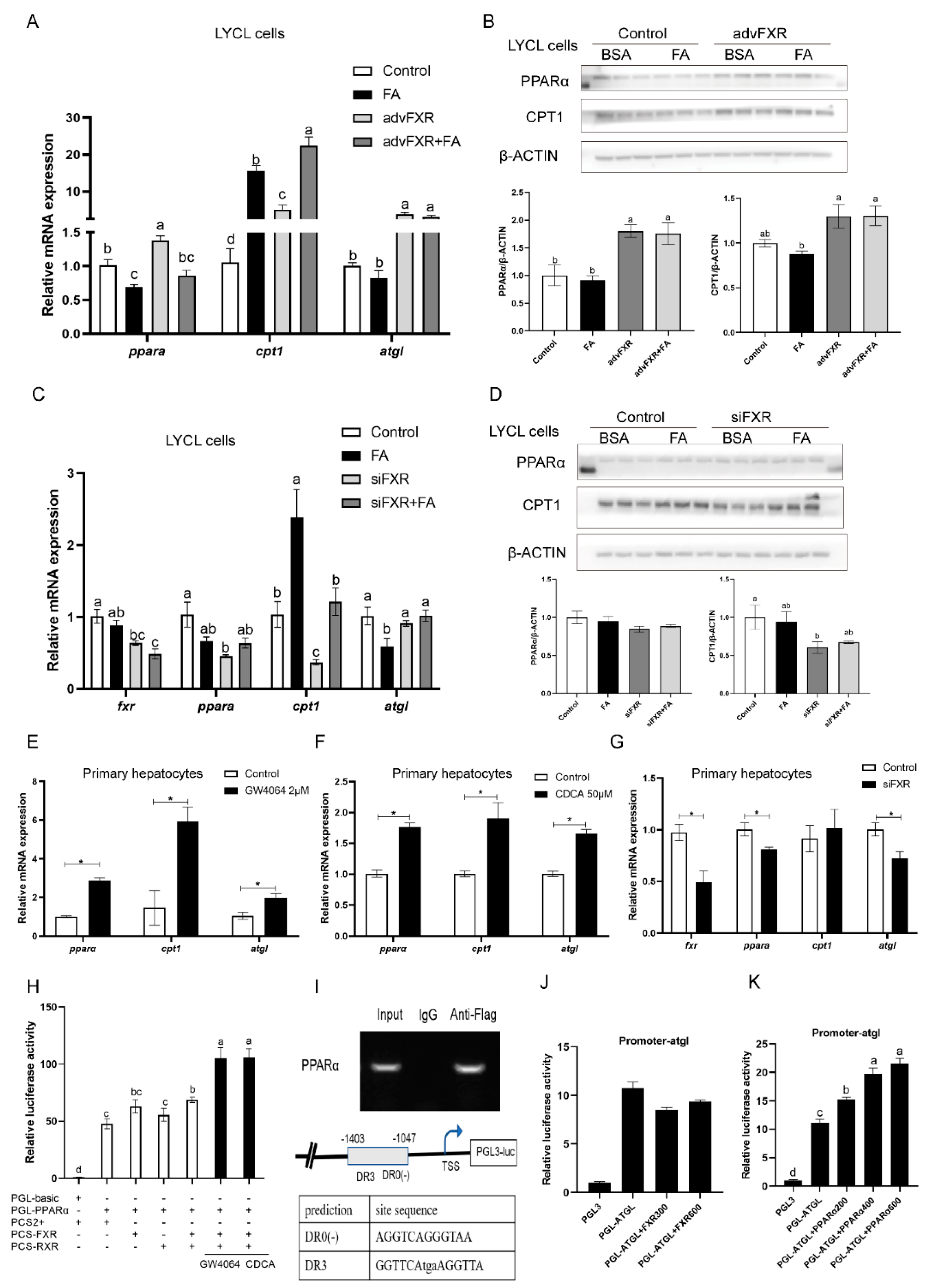
3.3. Overexpression and Agonists-Meditated Activation of FXR Decreases Triglycerides Concentration In Vitro
3.4. Regulation of FXR on Expression of pparα and Other Lipid Catabolism-Related Genes
3.5. Regulation of FXR on Expression of srebp1 and Other Lipogenesis Related Genes
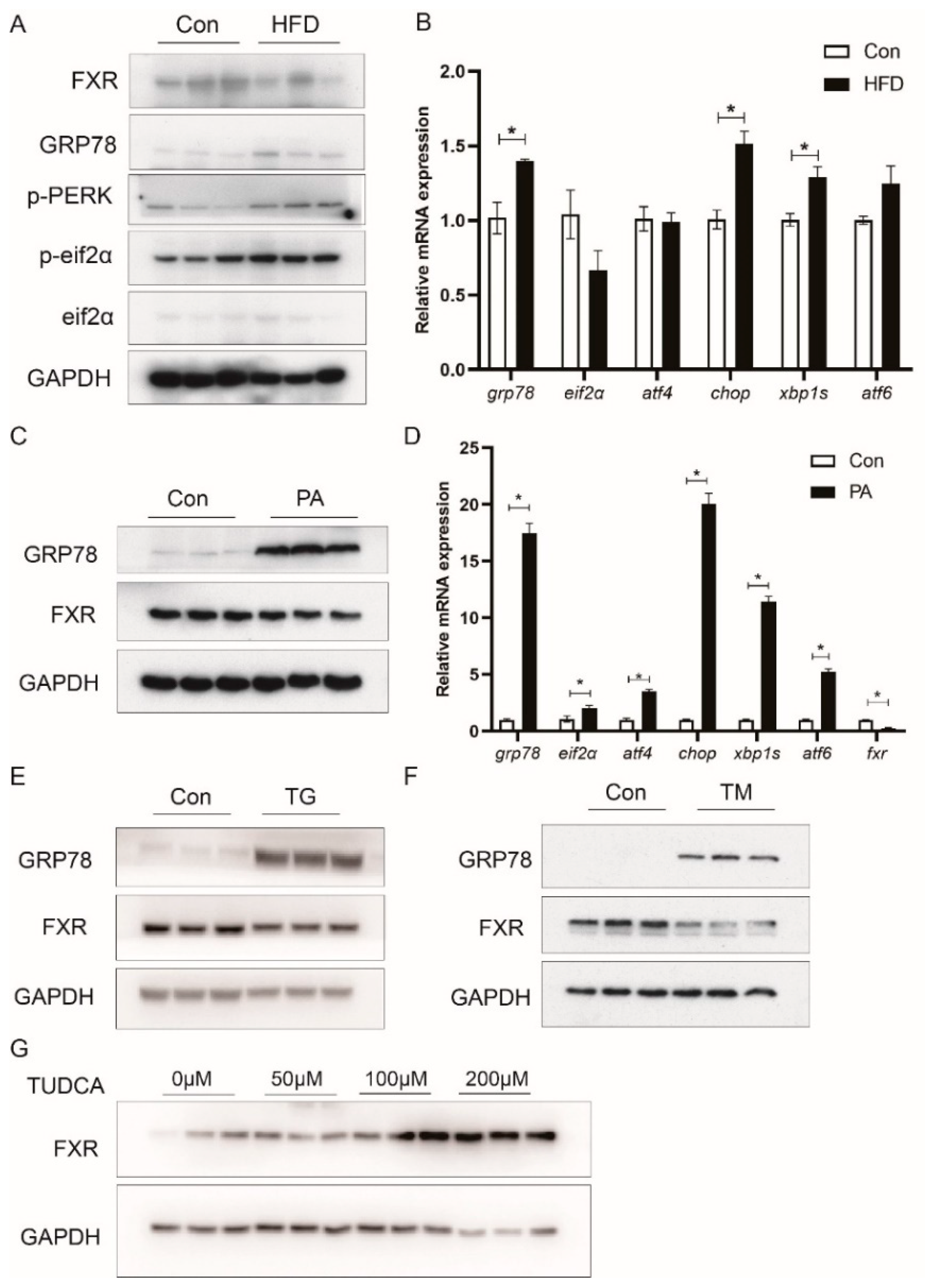
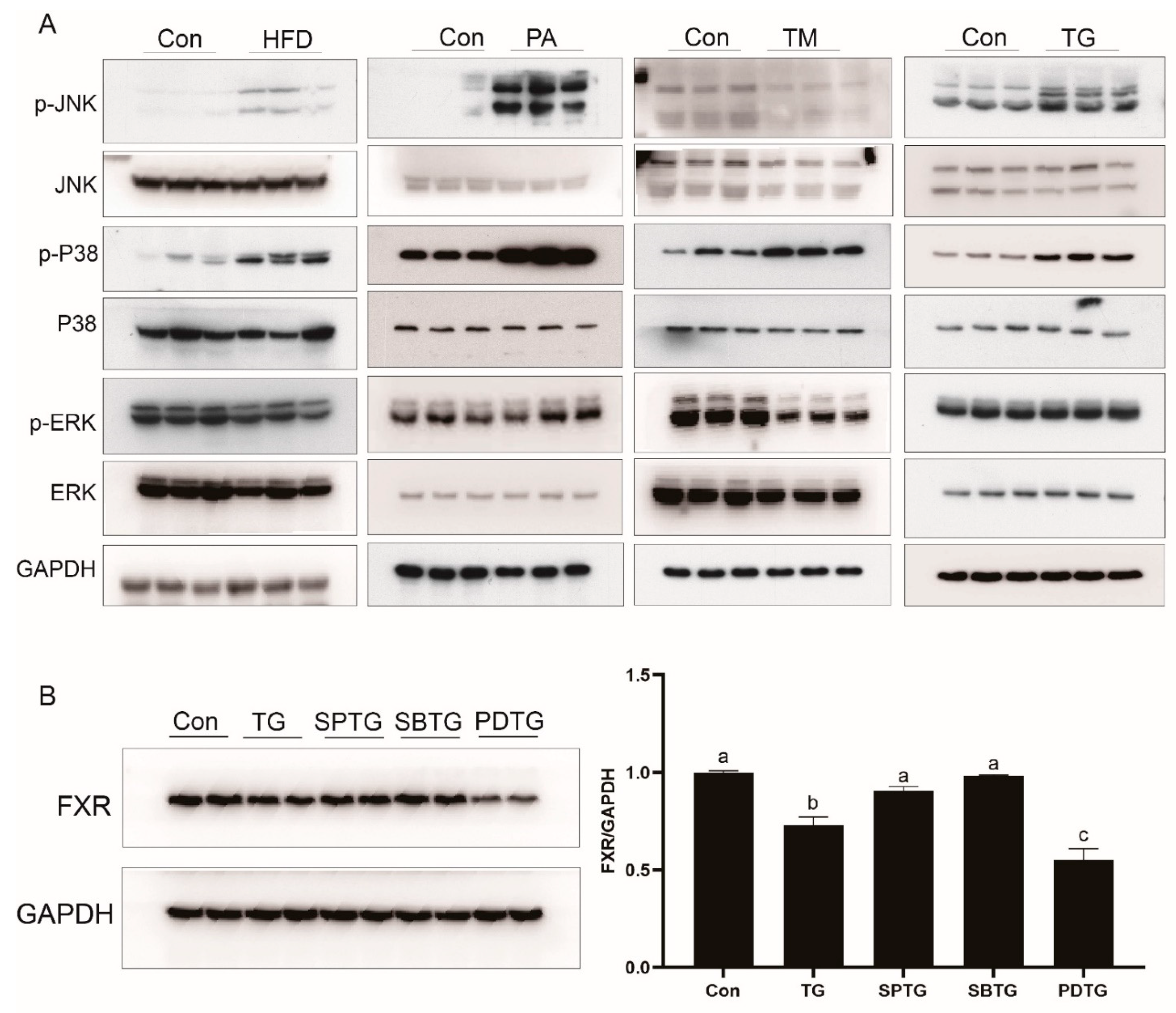
3.6. HFD Reduces FXR Expression in Association with ER Stress
3.7. ER Stress Reduces FXR Expression through the MAPK Pathway
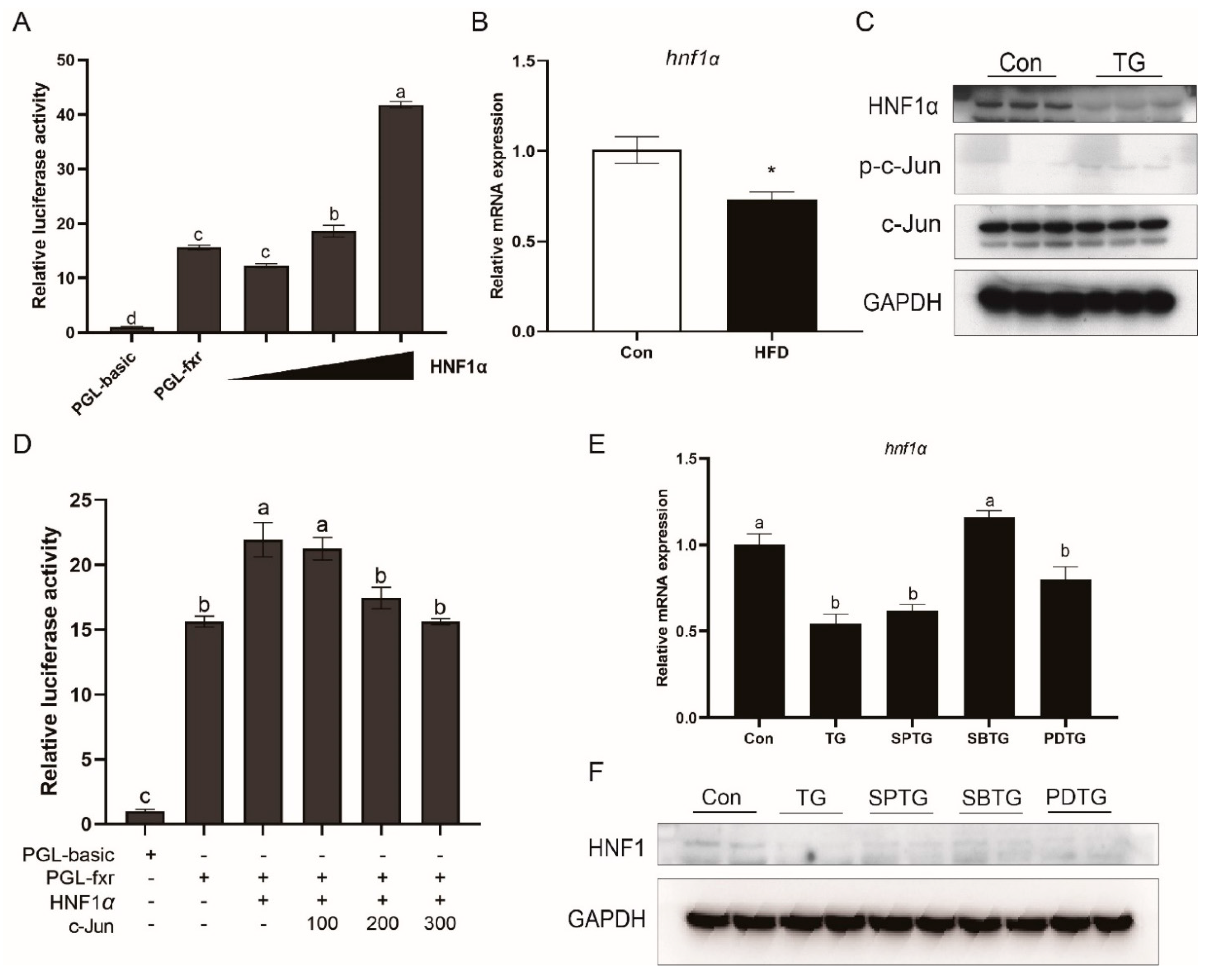
3.8. HNF1α Is Involved in Effects of ER Stress and MAPK Pathway on FXR
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Sanyal, A.J. NASH: A global health problem. Hepatol. Res. 2011, 41, 670–674. [Google Scholar] [CrossRef]
- Friedman, S.L.; Neuschwander-Tetri, B.A.; Rinella, M.; Sanyal, A.J. Mechanisms of NAFLD development and therapeutic strategies. Nat. Med. 2018, 24, 908–922. [Google Scholar] [CrossRef]
- Xi, Y.; Li, H. Role of farnesoid X receptor in hepatic steatosis in nonalcoholic fatty liver disease. Biomed. Pharmacother. 2020, 121, 109609. [Google Scholar] [CrossRef]
- Massafra, V.; van Mil, S. Farnesoid X receptor: A “homeostat” for hepatic nutrient metabolism. Biochim. Biophys. Acta Mol. Basis Dis. 2018, 1864, 45–59. [Google Scholar] [CrossRef]
- Watanabe, M.; Houten, S.M.; Wang, L.; Moschetta, A.; Mangelsdorf, D.J.; Heyman, R.A.; Moore, D.D.; Auwerx, J. Bile acids lower triglyceride levels via a pathway involving FXR, SHP, and SREBP-1c. J. Clin. Investig. 2004, 113, 1408–1418. [Google Scholar] [CrossRef] [Green Version]
- Sinal, C.J.; Tohkin, M.; Miyata, M.; Ward, J.M.; Lambert, G.; Gonzalez, F.J. Targeted Disruption of the Nuclear Receptor FXR/BAR Impairs Bile Acid and Lipid Homeostasis. Cell 2000, 102, 731–744. [Google Scholar] [CrossRef] [Green Version]
- Ma, Y.; Huang, Y.; Yan, L.; Gao, M.; Liu, D. Synthetic FXR Agonist GW4064 Prevents Diet-Induced Hepatic Steatosis and Insulin Resistance. Pharm. Res. 2013, 30, 1447–1457. [Google Scholar] [CrossRef] [PubMed]
- Cipriani, S.; Mencarelli, A.; Palladino, G.; Fiorucci, S. FXR activation reverses insulin resistance and lipid abnormalities and protects against liver steatosis in Zucker (fa/fa) obese rats. J. Lipid Res. 2010, 51, 771–784. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Correia, J.; Massart, J.; de Boer, J.; Porsmyr-Palmertz, M.; Martínez-Redondo, V.; Agudelo, L.; Sinha, I.; Meierhofer, D.; Ribeiro, V.; Björnholm, M.; et al. Bioenergetic cues shift FXR splicing towards FXRα2 to modulate hepatic lipolysis and fatty acid metabolism. Mol. Metab. 2015, 4, 891–902. [Google Scholar] [CrossRef]
- Teruo, M.; Akira, H.; Tadashi, I.; Mutsumi, S.; Yasushi, M. The effect of FXR ligands on fatty liver model of cultured cells. FASEB J. 2011, 25, 534–535. [Google Scholar]
- Adorini, L.; Pruzanski, M.; Shapiro, D. Farnesoid X receptor targeting to treat nonalcoholic steatohepatitis. Drug Discov. Today 2012, 17, 988–997. [Google Scholar] [CrossRef] [PubMed]
- Howarth, D.L.; Hagey, L.R.; Law, S.H.; Ai, N.; Krasowski, M.D.; Ekins, S.; Moore, J.T.; Kollitz, E.M.; Hinton, D.E.; Kullman, S.W. Two farnesoid X receptor alpha isoforms in Japanese medaka (Oryzias latipes) are differentially activated in vitro. Aquat. Toxicol. 2010, 98, 245–255. [Google Scholar] [CrossRef] [Green Version]
- Du, J.; Xiang, X.; Li, Y.; Ji, R.; Xu, H.; Mai, K.; Ai, Q. Molecular cloning and characterization of farnesoid X receptor from large yellow croaker (Larimichthys crocea) and the effect of dietary CDCA on the expression of inflammatory genes in intestine and spleen. Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 2018, 216, 10–17. [Google Scholar] [CrossRef]
- Maglich, J.M.; Caravella, J.A.; Lambert, M.H.; Willson, T.M.; Moore, J.T.; Ramamurthy, L. The first completed genome sequence from a teleost fish (Fugu rubripes) adds significant diversity to the nuclear receptor superfamily. Nucleic Acids Res. 2003, 31, 4051–4058. [Google Scholar] [CrossRef] [Green Version]
- Cai, S.; Xiong, L.; Wray, C.; Ballatori, N.; Boyer, J. The farnesoid X receptor FXRalpha/NR1H4 acquired ligand specificity for bile salts late in vertebrate evolution. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2007, 293, R1400–R1409. [Google Scholar] [CrossRef]
- Wu, K.; Zhao, T.; Hogstrand, C.; Xu, Y.-C.; Ling, S.-C.; Chen, G.-H.; Luo, Z. FXR-mediated inhibition of autophagy contributes to FA-induced TG accumulation and accordingly reduces FA-induced lipotoxicity. Cell Commun. Signal. 2020, 18, 1–16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reschly, E.J.; Ai, N.; Ekins, S.; Welsh, W.J.; Hagey, L.R.; Hofmann, A.F.; Krasowski, M. Evolution of the bile salt nuclear receptor FXR in vertebrates *. J. Lipid Res. 2008, 49, 1577–1587. [Google Scholar] [CrossRef] [Green Version]
- Krasowski, M.; Ni, A.; Hagey, L.; Ekins, S. Evolution of promiscuous nuclear hormone receptors: LXR, FXR, VDR, PXR, and CAR. Mol. Cell. Endocrinol. 2011, 334, 39–48. [Google Scholar] [CrossRef] [Green Version]
- Howarth, D.L.; Law, S.H.; Law, J.M.; Mondon, J.; Kullman, S.W.; Hinton, D.E. Exposure to the synthetic FXR agonist GW4064 causes alterations in gene expression and sublethal hepatotoxicity in eleutheroembryo medaka (Oryzias latipes). Toxicol. Appl. Pharmacol. 2010, 243, 111–121. [Google Scholar] [CrossRef] [Green Version]
- Du, J.; Chen, Q.; Li, Y.; Xiang, X.; Xu, W.; Mai, K.; Ai, Q. Activation of the Farnesoid X Receptor (FXR) Suppresses Linoleic Ac-id-Induced Inflammation in the Large Yellow Croaker (Larimichthys crocea). J. Nutr. 2020, 150, 2469–2477. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.-X.; Shen, W.; Sun, H. Effects of nuclear receptor FXR on the regulation of liver lipid metabolism in patients with non-alcoholic fatty liver disease. Hepatol. Int. 2010, 4, 741–748. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liao, Z.; Sun, B.; Zhang, Q.; Jia, L.; Wei, Y.; Liang, M.; Xu, H. Dietary bile acids regulate the hepatic lipid homeostasis in tiger puffer fed normal or high-lipid diets. Aquaculture 2020, 519, 734935. [Google Scholar] [CrossRef]
- Nie, H.; Song, C.; Wang, D.; Cui, S.; Ren, T.; Cao, Z.; Liu, Q.; Chen, Z.; Chen, X.; Zhou, Y. MicroRNA-194 inhibition improves di-etary-induced non-alcoholic fatty liver disease in mice through targeting on FXR. Biochim. Biophys. Acta Mol. Basis Dis. 2017, 1863, 3087–3094. [Google Scholar] [CrossRef] [PubMed]
- Xiong, X.; Wang, X.; Lu, Y.; Wang, E.; Zhang, Z.; Yang, J.; Zhang, H.; Li, X. Hepatic steatosis exacerbated by endoplasmic reticulum stress-mediated downregulation of FXR in aging mice. J. Hepatol. 2014, 60, 847–854. [Google Scholar] [CrossRef] [PubMed]
- Cao, X.; Dai, Y.; Liu, M.; Yuan, X.; Wang, C.; Huang, Y.; Liu, W.; Jiang, G. High-fat diet induces aberrant hepatic lipid secretion in blunt snout bream by activating endoplasmic reticulum stress-associated IRE1/XBP1 pathway. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2019, 1864, 213–223. [Google Scholar] [CrossRef] [PubMed]
- Jia, R.; Cao, L.-P.; Du, J.-L.; He, Q.; Gu, Z.-Y.; Jeney, G.; Xu, P.; Yin, G.-J. Effects of High-Fat Diet on Steatosis, Endoplasmic Reticulum Stress and Autophagy in Liver of Tilapia (Oreochromis niloticus). Front. Mar. Sci. 2020, 7, 363. [Google Scholar] [CrossRef]
- Li, J.; Huang, J.; Li, J.-S.; Chen, H.; Huang, K.; Zheng, L. Accumulation of endoplasmic reticulum stress and lipogenesis in the liver through generational effects of high fat diets. J. Hepatol. 2012, 56, 900–907. [Google Scholar] [CrossRef]
- Ji, R.; Xu, X.; Xiang, X.; Zhu, S.; Li, Y.; Mai, K.; Ai, Q. Regulation of adiponectin on lipid metabolism in large yellow croaker (Larimichthys crocea). Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2020, 1865, 158711. [Google Scholar] [CrossRef]
- Du, J.; Xu, H.; Li, S.; Cai, Z.; Mai, K.; Ai, Q. Effects of dietary chenodeoxycholic acid on growth performance, body composition and related gene expression in large yellow croaker (Larimichthys crocea) fed diets with high replacement of fish oil with soybean oil. Aquaculture 2017, 479, 584–590. [Google Scholar] [CrossRef]
- Li, S.; Monroig, O.; Wang, T.; Yuan, Y.; Navarro, J.C.; Hontoria, F.; Liao, K.; Tocher, D.; Mai, K.; Xu, W.; et al. Functional characterization and differential nutritional regulation of putative Elovl5 and Elovl4 elongases in large yellow croaker (Larimichthys crocea). Sci. Rep. 2017, 7, 1–15. [Google Scholar] [CrossRef]
- Tan, P.; Dong, X.; Mai, K.; Xu, W.; Ai, Q. Vegetable oil induced inflammatory response by altering TLR-NF-κB signalling, macrophages infiltration and polarization in adipose tissue of large yellow croaker (Larimichthys crocea). Fish Shellfish. Immunol. 2016, 59, 398–405. [Google Scholar] [CrossRef]
- Chatzifotis, S.; Panagiotidou, M.; Papaioannou, N.; Pavlidis, M.; Nengas, I.; Mylonas, C. Effect of dietary lipid levels on growth, feed utilization, body composition and serum metabolites of meagre (Argyrosomus regius) juveniles. Aquaculture 2010, 307, 65–70. [Google Scholar] [CrossRef]
- Yin, P.; Xie, S.; Zhuang, Z.; He, X.; Tang, X.; Tian, L.; Liu, Y.; Niu, J. Dietary supplementation of bile acid attenuate adverse effects of high-fat diet on growth performance, antioxidant ability, lipid accumulation and intestinal health in juvenile largemouth bass (Micropterus salmoides). Aquaculture 2021, 531, 735864. [Google Scholar] [CrossRef]
- Wang, J.-T.; Liu, Y.-J.; Tian, L.-X.; Mai, K.-S.; Du, Z.-Y.; Wang, Y.; Yang, H.-J. Effect of dietary lipid level on growth performance, lipid deposition, hepatic lipogenesis in juvenile cobia (Rachycentron canadum). Aquaculture 2005, 249, 439–447. [Google Scholar] [CrossRef]
- Han, T.; Li, X.; Wang, J.; Hu, S.; Jiang, Y.; Zhong, X. Effect of dietary lipid level on growth, feed utilization and body composition of juvenile giant croaker Nibea japonica. Aquaculture 2014, 434, 145–150. [Google Scholar] [CrossRef]
- Carr, R.M.; Reid, A.E. FXR Agonists as Therapeutic Agents for Non-alcoholic Fatty Liver Disease. Curr. Atheroscler. Rep. 2015, 17, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Evans, M.; Mahaney, P.; Marcucci, L.; Lai, K.; Wang, S.; Krueger, J.; Gardell, S.; Huard, C.; Martinez, R.; Vlasuk, G.; et al. A synthetic farnesoid X receptor (FXR) agonist promotes cholesterol lowering in models of dyslipidemia. Am. J. Physiol. Gastrointest. Liver Physiol. 2009, 296, G543–G552. [Google Scholar] [CrossRef] [Green Version]
- Zhang, D.-Y.; Zhu, L.; Liu, H.-N.; Tseng, Y.-J.; Weng, S.-Q.; Liu, T.-T.; Dong, L.; Shen, X.-Z. The protective effect and mechanism of the FXR agonist obeticholic acid via targeting gut microbiota in non-alcoholic fatty liver disease. Drug Des. Dev. Ther. 2019, ume 13, 2249–2270. [Google Scholar] [CrossRef] [Green Version]
- Ma, K.; Saha, P.K.; Chan, L.; Moore, D.D. Farnesoid X receptor is essential for normal glucose homeostasis. J. Clin. Investig. 2006, 116, 1102–1109. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pineda Torra, I.; Claudel, T.; Duval, C.; Kosykh, V.; Fruchart, J.-C.; Staels, B. Bile Acids Induce the Expression of the Human Peroxisome Proliferator-Activated Receptor Alpha Gene via Activation of the Farnesoid X Receptor. Mol. Endocrinol. 2003, 17, 259–272. [Google Scholar] [CrossRef] [Green Version]
- Varga, T.; Czimmerer, Z.; Nagy, L. PPARs are a unique set of fatty acid regulated transcription factors controlling both lipid metabolism and inflammation. Biochim. Biophys. Acta Mol. Basis Dis. 2011, 1812, 1007–1022. [Google Scholar] [CrossRef] [PubMed]
- Ning, L.-J.; He, A.-Y.; Li, J.-M.; Lu, D.-L.; Jiao, J.-G.; Li, L.-Y.; Zhang, M.-L.; Chen, L.-Q.; Du, Z.-Y. Mechanisms and metabolic regulation of PPARα activation in Nile tilapia (Oreochromis niloticus). Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2016, 1861, 1036–1048. [Google Scholar] [CrossRef]
- Sathyanarayan, A.; Mashek, M.T.; Mashek, D. ATGL Promotes Autophagy/Lipophagy via SIRT1 to Control Hepatic Lipid Droplet Catabolism. Cell Rep. 2017, 19, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Han, C.Y.; Rho, H.S.; Kim, A.; Kim, T.H.; Jang, K.; Jun, D.W.; Kim, J.W.; Kim, B.; Kim, S.G. FXR Inhibits Endoplasmic Reticulum Stress-Induced NLRP3 Inflammasome in Hepatocytes and Ameliorates Liver Injury. Cell Rep. 2018, 24, 2985–2999. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shih, D.Q.; Bussen, M.; Sehayek, E.; Ananthanarayanan, M.; Shneider, B.L.; Suchy, F.J.; Shefer, S.; Bollileni, J.S.; Gonzalez, F.J.; Breslow, J.L.; et al. Hepatocyte nuclear factor-1α is an essential regulator of bile acid and plasma cholesterol metabolism. Nat. Genet. 2001, 27, 375–382. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Du, J.; Xiang, X.; Xu, D.; Zhang, J.; Fang, W.; Xu, W.; Mai, K.; Ai, Q. FXR, a Key Regulator of Lipid Metabolism, Is Inhibited by ER Stress-Mediated Activation of JNK and p38 MAPK in Large Yellow Croakers (Larimichthys crocea) Fed High Fat Diets. Nutrients 2021, 13, 4343. https://0-doi-org.brum.beds.ac.uk/10.3390/nu13124343
Du J, Xiang X, Xu D, Zhang J, Fang W, Xu W, Mai K, Ai Q. FXR, a Key Regulator of Lipid Metabolism, Is Inhibited by ER Stress-Mediated Activation of JNK and p38 MAPK in Large Yellow Croakers (Larimichthys crocea) Fed High Fat Diets. Nutrients. 2021; 13(12):4343. https://0-doi-org.brum.beds.ac.uk/10.3390/nu13124343
Chicago/Turabian StyleDu, Jianlong, Xiaojun Xiang, Dan Xu, Junzhi Zhang, Wei Fang, Wei Xu, Kangsen Mai, and Qinghui Ai. 2021. "FXR, a Key Regulator of Lipid Metabolism, Is Inhibited by ER Stress-Mediated Activation of JNK and p38 MAPK in Large Yellow Croakers (Larimichthys crocea) Fed High Fat Diets" Nutrients 13, no. 12: 4343. https://0-doi-org.brum.beds.ac.uk/10.3390/nu13124343




