Administration with Quinoa Protein Reduces the Blood Pressure in Spontaneously Hypertensive Rats and Modifies the Fecal Microbiota
Abstract
:1. Introduction
2. Materials and Methods
2.1. Preparation of Quinoa Protein
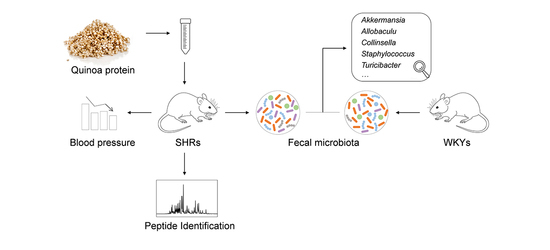
2.2. Animals and Experimental Design
2.3. Measurement of Blood Pressure
2.4. Collection of Fecal Samples
2.5. Collection of Digested Samples
2.6. LC–ESI–Q–TOF–MS Analysis
2.7. Gut Microbiota Analysis
2.8. Statistical Analysis
3. Results and Discussion
3.1. Identification of Peptides in Digested Samples
3.2. Antihypertensive Effect of Quinoa Protein on SHRs
3.3. Microbial Taxonomic Compositions of SHRs and WKYs
3.4. Improvement of Gut Microbial Dysbiosis in Treated SHRs
3.5. Key Genera Related to Blood Pressure in Rats
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Williams, B.; Mancia, G.; Spiering, W.; Agabiti Rosei, E.; Azizi, M.; Burnier, M.; Clement, D.L.; Coca, A.; de Simone, G.; Dominiczak, A.; et al. ESC/ESH guidelines for the management of arterial hypertension. Eur. Heart J. 2018, 39, 3021–3104. [Google Scholar] [CrossRef]
- Zhao, D.; Qi, Y.; Zheng, Z.; Wang, Y.; Zhang, X.Y.; Li, H.J.; Liu, H.H.; Zhang, X.T.; Du, J.; Liu, J. Dietary factors associated with hypertension. Nat. Rev. Cardiol. 2011, 8, 456–465. [Google Scholar] [CrossRef]
- Vasdev, S.; Stuckless, J. Antihypertensive effects of dietary protein and its mechanism. Int. J. Angiol. 2010, 19, e7–e20. [Google Scholar] [CrossRef] [Green Version]
- Dasinger, J.H.; Fehrenbach, D.J.; Abais-Battad, J.M. Dietary protein: Mechanisms influencing hypertension and renal disease. Curr. Hypertens. Rep. 2020, 22, 13. [Google Scholar] [CrossRef]
- Ait-Yahia, D.; Madani, S.; Savelli, J.L.; Prost, J.; Bouchenak, M.; Belleville, J. Dietary fish protein lowers blood pressure and alters tissue polyunsaturated fatty acid composition in spontaneously hypertensive rats. Nutrition 2003, 19, 342–346. [Google Scholar] [CrossRef]
- Pal, S.; Ellis, V. The chronic effects of whey proteins on blood pressure, vascular function, and inflammatory markers in overweight individuals. Obesity 2010, 18, 1354–1359. [Google Scholar] [CrossRef]
- de Oliveira, M.R.; Silva, T.J.; Barros, E.; Guimarães, V.M.; Baracat-Pereira, M.C.; Eller, M.R.; Reis Coimbra, J.S.; de Oliveira, E.B. Anti-Hypertensive Peptides Derived from Caseins: Mechanism of Physiological Action, Production Bioprocesses, and Challenges for Food Applications. Appl. Biochem. Biotechnol. 2018, 185, 884–908. [Google Scholar] [CrossRef]
- Rai, A.K.; Sanjukta, S.; Jeyaram, K. Production of angiotensin I converting enzyme inhibitory (ACE-I) peptides during milk fermentation and their role in reducing hypertension. Crit. Rev. Food Sci. 2017, 57, 2789–2800. [Google Scholar] [CrossRef] [PubMed]
- Richter, C.K.; Skulas-Ray, A.C.; Champagne, C.M.; Kris-Etherton, P.M. Plant protein and animal proteins: Do they differentially affect cardiovascular disease risk? Adv. Nutr. 2015, 6, 712–728. [Google Scholar] [CrossRef] [PubMed]
- Norris, R.; FitzGerald, R.J. Antihypertensive peptides from food proteins. In Bioactive Food Peptides in Health and Disease; Hernandez-Ledesma, B., Hsieh, C.C., Eds.; InTech Publishers: London, UK, 2013; pp. 45–72. [Google Scholar]
- Miralles, B.; Amigo, L.; Recio, I. Critical review and perspectives on food derived antihypertensive peptides. J. Agric. Food Chem. 2018, 66, 9384–9390. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.Y.; Hur, S.J. Antihypertensive peptides from animal products, marine organisms, and plants. Food Chem. 2017, 228, 506–517. [Google Scholar] [CrossRef] [PubMed]
- Adnan, S.; Nelson, J.W.; Ajami, N.J.; Venna, V.R.; Petrosino, J.F.; Bryan, R.M., Jr.; Durgan, D.J. Alterations in the gut microbiota can elicit hypertension in rats. Physiol. Genom. 2017, 49, 96–104. [Google Scholar] [CrossRef] [Green Version]
- Marques, F.Z.; Mackay, C.R.; Kaye, D.M. Beyond gut feelings: How the gut microbiota regulates blood pressure. Nat. Rev. Cardiol. 2018, 15, 20–32. [Google Scholar] [CrossRef]
- Tain, Y.L.; Lee, W.C.; Wu, K.L.H.; Leu, S.; Chan, J.Y.H. Resveratrol prevents the development of hypertension programmed by maternal plus post-weaning high-fructose consumption through modulation of oxidative stress, nutrient-sensing signals, and gut microbiota. Mol. Nutr. Food Res. 2018, 62, 1800066. [Google Scholar] [CrossRef]
- Yang, T.; Aquino, V.; Lobaton, G.O.; Li, H.B.; Colon-Perez, L.; Goel, R.; Qi, Y.F.; Zubcevic, J.; Febo, M.; Richards, E.M.; et al. Sustained captopril-induced reduction in blood pressure is associated with alterations in gut-brain axis in the spontaneously hypertensive rat. J. Am. Heart Assoc. 2019, 8, e010721. [Google Scholar] [CrossRef]
- Jama, H.A.; Beale, A.; Shihata, W.A.; Marques, F.Z. The effect of diet on hypertensive pathology: Is there a link via gut microbiota-driven immunometabolism? Cardiovasc. Res. 2019, 115, 1435–1447. [Google Scholar] [CrossRef]
- Wu, G.D.; Chen, J.; Hoffmann, C.; Bittinger, K.; Chen, Y.Y.; Keilbaugh, S.A.; Bewtra, M.; Knights, D.; Walters, W.A.; Knight, R.; et al. Linking long-term dietary patterns with gut microbial enterotypes. Science 2011, 334, 105–108. [Google Scholar] [CrossRef] [Green Version]
- Aluko, R.E.; Monu, E. Functional and bioactive properties of quinoa seed protein hydrolysates. J. Food Sci. 2003, 68, 1254–1258. [Google Scholar] [CrossRef]
- Mudgil, P.; Kilari, B.P.; Kamal, H.; Olalere, O.A.; FitzGerald, R.J.; Gan, C.H.; Maqsood, S. Multifunctional bioactive peptides derived from quinoa protein hydrolysates: Inhibition of α-glucosidase, dipeptidyl peptidase-IV and angiotensin I converting enzymes. J. Cereal Sci. 2020, 96, 103130. [Google Scholar] [CrossRef]
- Ujiroghene, O.J.; Liu, L.; Zhang, S.W.; Lu, J.; Pang, X.Y.; Lv, J.P. α-Glucosidase and ACE dual inhibitory protein hydrolysates and peptide fractions of sprouted quinoa yoghurt beverages inoculated with Lactobacillus casei. Food Chem. 2019, 299, 124985. [Google Scholar]
- Zheng, Y.J.; Wang, X.; Zhuang, Y.L.; Li, Y.; Tian, H.L.; Shi, P.Q.; Li, G.F. Isolation of novel ACE-inhibitory and antioxidant peptides from quinoa bran albumin assisted with an in silico approach: Characterization, in vivo antihypertension, and molecular docking. Molecules 2019, 24, 4562. [Google Scholar] [CrossRef] [Green Version]
- Guo, H.M.; Hao, Y.Q.; Richel, A.; Everaert, N.; Chen, Y.H.; Liu, M.J.; Yang, X.S.; Ren, G.X. Antihypertensive effect of quinoa protein under simulated gastrointestinal digestion and peptide characterization. J. Sci. Food Agric. 2020, 100, 5569–5576. [Google Scholar] [CrossRef]
- Guo, H.M.; Richel, A.; Hao, Y.Q.; Everaert, N.; Yang, X.S.; Ren, G.X. Novel dipeptidyl peptidase-IV and angiotensin-I-converting enzyme inhibitory peptides released from quinoa protein by in silico proteolysis. Food Sci. Nutr. 2020, 8, 1415–1422. [Google Scholar] [CrossRef] [Green Version]
- Fritz, M.; Rinaldi, G. Blood pressure measurement with the tail-cuff method in Wistar and spontaneously hypertensive rats: Influence of adrenergic- and nitric oxide-mediated vasomotion. J. Pharmacol. Toxicol. Methods 2008, 58, 215–221. [Google Scholar] [CrossRef] [PubMed]
- Boutrou, R.; Gaudichon, C.; Dupont, D.; Jardin, J.; Airinei, G.; Marsset-Baglieri, A.; Benamouzig, R.; Tome, D.; Leonil, J. Sequential release of milk protein-derived bioactive peptides in the jejunum in healthy humans. Am. J. Clin. Nutr. 2013, 97, 1314–1323. [Google Scholar] [CrossRef] [Green Version]
- Boutrou, R.; Henry, G.; Sanchez-Rivera, L. On the trail of milk bioactive peptides in human and animal intestinal tracts during digestion: A review. Dairy Sci. Technol. 2015, 95, 815–829. [Google Scholar] [CrossRef] [Green Version]
- Portune, K.J.; Beaumont, M.; Davila, A.M.; Tomé, D.; Blachier, F.; Sanz, Y. Gut microbiota role in dietary protein metabolism and health-related outcomes: The two sides of the coin. Trends Food Sci. Technol. 2016, 57, 213–232. [Google Scholar] [CrossRef] [Green Version]
- Davila, A.M.; Blachier, F.; Gotteland, M.; Andriamihaja, M.; Benetti, P.H.; Sanz, Y.; Tomé, D. Intestinal luminal nitrogen metabolism: Role of the gut microbiota and consequences for the host. Pharmacol. Res. 2013, 68, 95–107. [Google Scholar] [CrossRef]
- Barbe, F.; Le Feunteun, S.; Remond, D.; Ménard, O.; Jardin, J.; Henry, G.; Laroche, B.; Dupont, D. Tracking the in vivo release of bioactive peptides in the gut during digestion: Mass spectrometry peptidomic characterization of effluents collected in the gut of dairy matrix. Food Res. Int. 2014, 63, 147–156. [Google Scholar] [CrossRef]
- Volpe, M.; Gallo, G.; Tocci, G. Is early and fast blood pressure control important in hypertension management? Int. J. Cardiol. 2018, 254, 328–332. [Google Scholar] [CrossRef]
- Gupta, R.; Guptha, S. Strategies for initial management of hypertension. Indian J. Med. Res. 2010, 132, 531–542. [Google Scholar]
- Mell, B.; Jala, V.R.; Mathew, A.V.; Byun, J.; Waghulde, H.; Zhang, Y.; Haribabu, B.; Vijay-Kumar, M.; Pennathur, S.; Joe, B. Evidence for a link between gut microbiota and hypertension in the Dahl rat. Physiol. Genom. 2015, 47, 187–197. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Durgan, D.J.; Ganesh, B.P.; Cope, J.L.; Ajami, N.J.; Phillips, S.C.; Petrosino, J.F.; Hollister, E.B.; Bryan, R.M. Role of the gut microbiome in obstructive sleep apnea-induced hypertension. Hypertension 2016, 67, 469–474. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, T.; Santisteban, M.M.; Rodriguez, V.; Li, E.; Ahmari, N.; Marulanda Carvajal, J.; Zadeh, M.; Gong, M.; Qi, Y.; Zubcevic, J.; et al. Gut dysbiosis is linked to hypertension. Hypertension 2015, 65, 1331–1340. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoffman, J.D.; Parikh, I.; Green, S.J.; Chlipala, G.; Mohney, R.P.; Keaton, M.; Bauer, B.; Hartz, A.M.S.; Lin, A.L. Age drives distortion of brain metabolic, vascular and cognitive functions, and the gut microbiome. Front. Aging Neurosci. 2017, 9, 298. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Badal, V.D.; Vaccariello, E.D.; Murray, E.R.; Yu, K.E.; Knight, R.; Jeste, D.V.; Nguyen, T.T. The gut microbiome, aging, and longevity: A systematic review. Nutrients 2020, 12, 3759. [Google Scholar] [CrossRef]
- Lopetuso, L.R.; Quagliariello, A.; Schiavoni, M.; Petito, V.; Russo, A.; Reddel, S.; Chierico, F.D.; Ianiro, G.; Scaldaferri, F.; Neri, M.; et al. Towards a disease-associated common trait of gut microbiota dysbiosis: The pivotal role of Akkermansia muciniphila. Digest. Liver Dis. 2020, 52, 1002–1010. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Bei, J.; Liang, L.; Yu, G.; Li, L.; Li, Q. Stachyose improves inflammation through modulating gut microbiota of high-fat diet/streptozotocin-induced type 2 diabetes in rats. Mol. Nutr. Food Res. 2018, 62, 1700954. [Google Scholar] [CrossRef]
- Chen, L.; Chen, D.Q.; Liu, J.R.; Zhang, J.; Vaziri, N.D.; Zhuang, S.; Chen, H.; Feng, Y.L.; Guo, Y.; Zhao, Y.Y. Unilateral ureteral obstruction causes gut microbial dysbiosis and metabolome disorders contributing to tubulointerstitial fibrosis. Exp. Mol. Med. 2019, 51, 38. [Google Scholar] [CrossRef] [Green Version]
- Gu, Z.; Zhu, Y.; Jiang, S.; Xia, G.; Li, C.; Zhang, X.; Zhang, J.; Shen, X. Tilapia head glycolipids reduce inflammation by regulating the gut microbiota in dextran sulphate sodium-induced colitis mice. Food Funct. 2020, 11, 3245–3255. [Google Scholar] [CrossRef]
- Presley, L.L.; Wei, B.; Braun, J.; Borneman, J. Bacteria associated with immunoregulatory cells in mice. Appl. Environ. Microbiol. 2010, 76, 936–941. [Google Scholar] [CrossRef] [Green Version]
- Zhong, Y.; Nyman, M.; Fak, F. Modulation of gut microbiota in rats fed high-fat diets by processing whole-grain barley to barley malt. Mol. Nutr. Food Res. 2015, 59, 2066–2076. [Google Scholar] [CrossRef] [PubMed]
- Naqvi, S.; Asar, T.O.; Kumar, V.; Al-Abbasi, F.A.; Alhayyani, S.; Kamal, M.A.; Anwar, F. A cross-talk between gut microbiome, salt and hypertension. Biomed. Pharmacother. 2021, 134, 111156. [Google Scholar] [CrossRef] [PubMed]
- Metzler-Zebeli, B.U.; Mann, E.; Schmitz-Esser, S.; Wagner, M.; Ritzmann, M.; Zebelib, Q. Changing dietary calcium-phosphorus level and cereal source selectively alters abundance of bacteria and metabolites in the upper gastrointestinal tracts of weaned pigs. Appl. Environ. Microbiol. 2013, 79, 7264–7272. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Al Khodor, S.; Reichert, B.; Shatat, I.F. The microbiome and blood pressure: Can microbes regulate our blood pressure? Front. Pediatr. 2017, 5, 138. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.S.; Davies, S.S. Microbial metabolism of dietary components to bioactive metabolites: Opportunities for new therapeutic interventions. Genome Med. 2016, 8, 46. [Google Scholar] [CrossRef] [PubMed] [Green Version]




| Quinoa Protein | ACE Inhibitory Peptides |
|---|---|
| 13S globulin seed storage protein 2-like: MSRVFLLPLALTLLILSPTSLAQLGFQLGQSPFLPSGQSSPQHSRLQRGQQALNDCQINQLSANEPSIRIQAEAGITEVWDPKEQQEFQCAGVTVIRREIEPKGLLLPHYNNAPSISYVIRGRGLLGLSSLGCADTYESGSPEFFSEESRRSERFEESRRSERGSEEMRDQHQKVRRFHKGHVIGLPAGVSKWVYNDGEDRLTIVTLYDTNNFQNQLDDNLRSFFLAGNPQGRGGDQSGRQHESSRRHTRGGQEEMGQNILSGFDKQLLADAFEVESDTISKIQGENDDRGAIIRVESGELEMLIPEWDQEEQRSERHHRGGGSERSEEEERSERHHRGGRGRQSESSRPHNGIEQTLCSARLSVNIDNPERADVFNPQGGRLTNINSNKLPILNYLRLSAEKVNLYKNAIMTPNWKINAHSIVYFTKGSGRVQIANHEGELVFDDMVQEGQLVVVPQNFVVLKRAGQDGLEWVALLTNDNAMSSPLAGRISAIRGMPIEVVMNSYKLSREEAQRLKYGRQELSVFSPSKRSERRGDEYAIV | LG, RL, LQ, GF, FQ, GQ, SG, PQ, LG, RL, LQIR, VW, KE, GL, LLP, PH, HY, YN, AP, NF, FQ, NLR, LR, SF, GG, GQ, MG, IL, SG, GF, FDK, EV, VE, QG, GE, IP, EW, EQR, ER, RA, NK, NKL, KLP, KL, IL, LN, LNY, NY, IA, EG, GE, LVF, VF, DM, PL, LA, AG, GR |
| legumin A-like: MAKSTTTLFLLSCSIALVLLNGCMGQGRMREMQGNECQIDRLTALEPTHRIQAEGGLTEVWDTQDQQFQCSGVSVIRRTIEPNGLLLPSFTSGPELIYIEQGNGISGLMIPGCPETFESMSQESWREGMERGMRGGRFQDQHQKIRHLRQGHIFAMPAGVAHWAYNSGNEPLVAVILIDTSNHANQLDKDYPKRFYLAGKPQQEHSRHHHRGGESQRGEHGSDGNVFSGLDTKSVAQSFGVSEDIAEKLQAKQDERGNIVLVQEGLHVIKPPSSRSYDDEREQRHRSPRSNGLEETICSARLSENIDDPSKADVYSPEAGRLTTLNSFNLPILSNLRLSAEKGVLYRNAIMAPHYNLNAHSIIYGVRGRGRIQIVNAQGNSVFDDELRQGQLVVVPQNFAVVKQAGEEGFEWIAFKTCENALFQTLAGRTSAIRAMPVEVISNIYQISREQAYRLKFSRSETTLFRPENQGRQRREMAA | ALEP, PT, AG, GV, AH, WA, AY, YN, SG, AG, GV, AH, WA, DY, YP, KR, RF, SF, SG, FG, GV, IA, IAE, EK, KL, LQ, GL, SF, SG, FG, GV, IA, IAE, EK, KL, LQ, GL, ER, RG, EG, GL, VIKP, IKP, KP, PP, KA, VYEA, AG, GR, RL, LN, SF, NLR, LR, RL, EK, KG, GV, GVLY, VLY, LY, AI, MAP, AP, PH, HY, YN, GR, VF, LR, GQ, QG, AV, EW, AVV, VK, AG, GE, EG, GF, EW, AI, IR, IRA, RA, LF, LFR, FR, RP, QG, GR |
| 11S globulin seed storage protein 2-like: MGGTKILVALSLCLMVSSALGQGSHKRLSYQAQQCRINRLTSSEPNQRIECEGGLIELWDETEEQFQCSGIHAMRVTVQQNSLSLPNFHPFPRLVYIERGEGILGVTFPGCPETYDSSGRQEERIRGDEQREFGQQKDLHQKVHRFTRGDIIAIPPGAVHWCYNDGNEEVVTVIVNDLNNPSNQLDQTFRSFYLAGGLEKSSEIRGKINNIFRPFAPELLSEAFDVPEDLIRKMQQTENRGLIIRVDKGEMRILSPGSEQDYDDERRRKYVGLDVNGLEETICTMRLRHNLDNRREADVYSRHGGRLNIVNEHKLPILRHLDMSVEKGNMFPNTIYSPHWAVNSHSVVYVTRGEAHIQVVGNNGESVMDDRVNEGEMFVIPQYFTVSVKAGSNGFEYVSFKTTSSPMKSPMVGYTSVLRAMPVQVLTNAYQISPSEAHQLKYNREHQTFFLPSRGGRSRRF | NF, HP, PFP, FP, PR, RG, GD, IAIPP, IA, IAIP, AIPP, AI, AIP, IPP, IP, PP, PG, GA, AV, LN, TF, FR, EI, IR, RG, GK, IF, FR, RP, IR, TE, RG, GL, IL, LSP, PG, GS, DY, ER, HG, GG, GR, RL, LN, HK, KL, KLP, LPILR, IL, LR, VE, EK, KG, MF, FP, IY, IYSPH, PH, WA, AV, VG, GE, NG, VM, EG, VK, KA, AG, AGS, GS, NG, GF, EY, YV, SF, VG, GY, EA, AH, KY, YN |
| PERMANOVA (Pairwise Test) | P | |
|---|---|---|
| Bray–Curtis | Unweighted UniFrac | |
| SHR–S vs SHR–Q | 0.003 | 0.012 |
| SHR–S vs SHR–C | 0.003 | 0.001 |
| SHR–S vs WKY–S | 0.001 | 0.001 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guo, H.; Hao, Y.; Fan, X.; Richel, A.; Everaert, N.; Yang, X.; Ren, G. Administration with Quinoa Protein Reduces the Blood Pressure in Spontaneously Hypertensive Rats and Modifies the Fecal Microbiota. Nutrients 2021, 13, 2446. https://0-doi-org.brum.beds.ac.uk/10.3390/nu13072446
Guo H, Hao Y, Fan X, Richel A, Everaert N, Yang X, Ren G. Administration with Quinoa Protein Reduces the Blood Pressure in Spontaneously Hypertensive Rats and Modifies the Fecal Microbiota. Nutrients. 2021; 13(7):2446. https://0-doi-org.brum.beds.ac.uk/10.3390/nu13072446
Chicago/Turabian StyleGuo, Huimin, Yuqiong Hao, Xin Fan, Aurore Richel, Nadia Everaert, Xiushi Yang, and Guixing Ren. 2021. "Administration with Quinoa Protein Reduces the Blood Pressure in Spontaneously Hypertensive Rats and Modifies the Fecal Microbiota" Nutrients 13, no. 7: 2446. https://0-doi-org.brum.beds.ac.uk/10.3390/nu13072446