Minimal Effects of Medium-Chain Triglyceride Supplementation on the Intestinal Microbiome Composition of Premature Infants: A Single-Center Pilot Study
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Participants and Eligibility
2.2. Sample Collection
2.3. DNA Extraction, 16s rRNA Amplification, Library Preparation, and Sequencing
2.4. Microbiota Analysis
2.5. Statistical Analyses
3. Results
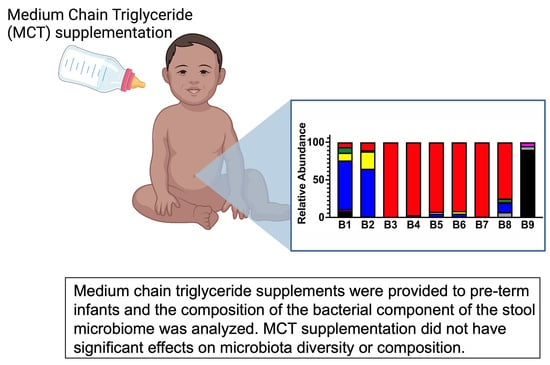
3.1. Pre-Term Infants Harbor a Limited Gut Microbiota Composition Dominated by Enterobacteriaceae and Not Impacted by MCT Supplementation
3.2. Human Milk vs. Formula Feeding
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Backhed, F.; Roswall, J.; Peng, Y.; Feng, Q.; Jia, H.; Kovatcheva-Datchary, P.; Li, Y.; Xia, Y.; Xie, H.; Zhong, H.; et al. Dynamics and Stabilization of the Human Gut Microbiome during the First Year of Life. Cell Host Microbe 2015, 17, 690–703. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dominguez-Bello, M.G.; Costello, E.K.; Contreras, M.; Magris, M.; Hidalgo, G.; Fierer, N.; Knight, R. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc. Natl. Acad. Sci. USA 2010, 107, 11971–11975. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shao, Y.; Forster, S.C.; Tsaliki, E.; Vervier, K.; Strang, A.; Simpson, N.; Kumar, N.; Stares, M.D.; Rodger, A.; Brocklehurst, P.; et al. Stunted microbiota and opportunistic pathogen colonization in caesarean-section birth. Nature 2019, 574, 117–121. [Google Scholar] [CrossRef] [PubMed]
- Ferretti, P.; Pasolli, E.; Tett, A.; Asnicar, F.; Gorfer, V.; Fedi, S.; Armanini, F.; Truong, D.T.; Manara, S.; Zolfo, M.; et al. Mother-to-Infant Microbial Transmission from Different Body Sites Shapes the Developing Infant Gut Microbiome. Cell Host Microbe 2018, 24, 133–145.e5. [Google Scholar] [CrossRef] [PubMed]
- Charbonneau, M.R.; Blanton, L.V.; DiGiulio, D.B.; Relman, D.A.; Lebrilla, C.B.; Mills, D.A.; Gordon, J.I. A microbial perspective of human developmental biology. Nature 2016, 535, 48–55. [Google Scholar] [CrossRef] [Green Version]
- Brooks, B.; Firek, B.A.; Miller, C.S.; Sharon, I.; Thomas, B.C.; Baker, R.; Morowitz, M.J.; Banfield, J.F. Microbes in the neonatal intensive care unit resemble those found in the gut of premature infants. Microbiome 2014, 2, 1. [Google Scholar] [CrossRef] [Green Version]
- Young, G.R.; van der Gast, C.J.; Smith, D.L.; Berrington, J.E.; Embleton, N.D.; Lanyon, C. Acquisition and Development of the Extremely Preterm Infant Microbiota Across Multiple Anatomical Sites. J. Pediatr. Gastroenterol. Nutr. 2020, 70, 12–19. [Google Scholar] [CrossRef] [Green Version]
- Azad, M.B.; Konya, T.; Maughan, H.; Guttman, D.S.; Field, C.J.; Chari, R.S.; Sears, M.R.; Becker, A.B.; Scott, J.A.; Kozyrskyj, A.L.; et al. Gut microbiota of healthy Canadian infants: Profiles by mode of delivery and infant diet at 4 months. CMAJ 2013, 185, 385–394. [Google Scholar] [CrossRef] [Green Version]
- Stewart, C.J.; Ajami, N.J.; O’Brien, J.L.; Hutchinson, D.S.; Smith, D.P.; Wong, M.C.; Ross, M.C.; Lloyd, R.E.; Doddapaneni, H.; Metcalf, G.A.; et al. Temporal development of the gut microbiome in early childhood from the TEDDY study. Nature 2018, 562, 583–588. [Google Scholar] [CrossRef]
- Martin, R.; Makino, H.; Cetinyurek Yavuz, A.; Ben-Amor, K.; Roelofs, M.; Ishikawa, E.; Kubota, H.; Swinkels, S.; Sakai, T.; Oishi, K.; et al. Early-Life Events, Including Mode of Delivery and Type of Feeding, Siblings and Gender, Shape the Developing Gut Microbiota. PLoS ONE 2016, 11, e0158498. [Google Scholar] [CrossRef] [Green Version]
- Sevelsted, A.; Stokholm, J.; Bonnelykke, K.; Bisgaard, H. Cesarean section and chronic immune disorders. Pediatrics 2015, 135, e92–e98. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mayer-Davis, E.J.; Rifas-Shiman, S.L.; Zhou, L.; Hu, F.B.; Colditz, G.A.; Gillman, M.W. Breast-feeding and risk for childhood obesity: Does maternal diabetes or obesity status matter? Diabetes Care 2006, 29, 2231–2237. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ihekweazu, F.D.; Versalovic, J. Development of the Pediatric Gut Microbiome: Impact on Health and Disease. Am. J. Med. Sci. 2018, 356, 413–423. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Milani, C.; Duranti, S.; Bottacini, F.; Casey, E.; Turroni, F.; Mahony, J.; Belzer, C.; Delgado Palacio, S.; Arboleya Montes, S.; Mancabelli, L.; et al. The First Microbial Colonizers of the Human Gut: Composition, Activities, and Health Implications of the Infant Gut Microbiota. Microbiol. Mol. Biol. Rev. 2017, 81, e00036-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arrieta, M.C.; Stiemsma, L.T.; Dimitriu, P.A.; Thorson, L.; Russell, S.; Yurist-Doutsch, S.; Kuzeljevic, B.; Gold, M.J.; Britton, H.M.; Lefebvre, D.L.; et al. Early infancy microbial and metabolic alterations affect risk of childhood asthma. Sci. Transl. Med. 2015, 7, 307ra152. [Google Scholar] [CrossRef]
- Stiemsma, L.T.; Turvey, S.E. Asthma and the microbiome: Defining the critical window in early life. Allergy Asthma Clin. Immunol. 2017, 13, 3. [Google Scholar] [CrossRef] [Green Version]
- Boutin, R.C.T.; Sbihi, H.; McLaughlin, R.J.; Hahn, A.S.; Konwar, K.M.; Loo, R.S.; Dai, D.; Petersen, C.; Brinkman, F.S.L.; Winsor, G.L.; et al. Composition and Associations of the Infant Gut Fungal Microbiota with Environmental Factors and Childhood Allergic Outcomes. mBio 2021, 12, e0339620. [Google Scholar] [CrossRef]
- Duffy, L.C. Interactions mediating bacterial translocation in the immature intestine. J. Nutr. 2000, 130, 432S–436S. [Google Scholar] [CrossRef]
- Halpern, M.D.; Denning, P.W. The role of intestinal epithelial barrier function in the development of NEC. Tissue Barriers 2015, 3, e1000707. [Google Scholar] [CrossRef] [Green Version]
- Tarr, P.I.; Warner, B.B. Gut bacteria and late-onset neonatal bloodstream infections in preterm infants. Semin. Fetal Neonatal Med. 2016, 21, 388–393. [Google Scholar] [CrossRef]
- Carl, M.A.; Ndao, I.M.; Springman, A.C.; Manning, S.D.; Johnson, J.R.; Johnston, B.D.; Burnham, C.A.; Weinstock, E.S.; Weinstock, G.M.; Wylie, T.N.; et al. Sepsis from the gut: The enteric habitat of bacteria that cause late-onset neonatal bloodstream infections. Clin. Infect. Dis. 2014, 58, 1211–1218. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sherman, M.P. New concepts of microbial translocation in the neonatal intestine: Mechanisms and prevention. Clin. Perinatol. 2010, 37, 565–579. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jacquot, A.; Neveu, D.; Aujoulat, F.; Mercier, G.; Marchandin, H.; Jumas-Bilak, E.; Picaud, J.C. Dynamics and clinical evolution of bacterial gut microflora in extremely premature patients. J. Pediatr. 2011, 158, 390–396. [Google Scholar] [CrossRef] [PubMed]
- Arboleya, S.; Binetti, A.; Salazar, N.; Fernandez, N.; Solis, G.; Hernandez-Barranco, A.; Margolles, A.; de Los Reyes-Gavilan, C.G.; Gueimonde, M. Establishment and development of intestinal microbiota in preterm neonates. FEMS Microbiol. Ecol. 2012, 79, 763–772. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Butel, M.J.; Suau, A.; Campeotto, F.; Magne, F.; Aires, J.; Ferraris, L.; Kalach, N.; Leroux, B.; Dupont, C. Conditions of bifidobacterial colonization in preterm infants: A prospective analysis. J. Pediatr. Gastroenterol. Nutr. 2007, 44, 577–582. [Google Scholar] [CrossRef]
- Madan, J.C.; Salari, R.C.; Saxena, D.; Davidson, L.; O’Toole, G.A.; Moore, J.H.; Sogin, M.L.; Foster, J.A.; Edwards, W.H.; Palumbo, P.; et al. Gut microbial colonisation in premature neonates predicts neonatal sepsis. Arch. Dis. Child. Fetal Neonatal Ed. 2012, 97, F456–F462. [Google Scholar] [CrossRef]
- Mai, V.; Torrazza, R.M.; Ukhanova, M.; Wang, X.; Sun, Y.; Li, N.; Shuster, J.; Sharma, R.; Hudak, M.L.; Neu, J. Distortions in development of intestinal microbiota associated with late onset sepsis in preterm infants. PLoS ONE 2013, 8, e52876. [Google Scholar] [CrossRef]
- Zhou, Y.; Shan, G.; Sodergren, E.; Weinstock, G.; Walker, W.A.; Gregory, K.E. Longitudinal analysis of the premature infant intestinal microbiome prior to necrotizing enterocolitis: A case-control study. PLoS ONE 2015, 10, e0118632. [Google Scholar] [CrossRef]
- Saiman, L.; Ludington, E.; Pfaller, M.; Rangel-Frausto, S.; Wiblin, R.T.; Dawson, J.; Blumberg, H.M.; Patterson, J.E.; Rinaldi, M.; Edwards, J.E.; et al. Risk factors for candidemia in Neonatal Intensive Care Unit patients. The National Epidemiology of Mycosis Survey study group. Pediatr. Infect. Dis. J. 2000, 19, 319–324. [Google Scholar] [CrossRef]
- Qin, J.; Li, R.; Raes, J.; Arumugam, M.; Burgdorf, K.S.; Manichanh, C.; Nielsen, T.; Pons, N.; Levenez, F.; Yamada, T.; et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 2010, 464, 59–65. [Google Scholar] [CrossRef] [Green Version]
- Stoll, B.J.; Hansen, N.; Fanaroff, A.A.; Wright, L.L.; Carlo, W.A.; Ehrenkranz, R.A.; Lemons, J.A.; Donovan, E.F.; Stark, A.R.; Tyson, J.E.; et al. Late-onset sepsis in very low birth weight neonates: The experience of the NICHD Neonatal Research Network. Pediatrics 2002, 110, 285–291. [Google Scholar] [CrossRef] [PubMed]
- Russell, C.; Lay, K.M. Natural history of Candida species and yeasts in the oral cavities of infants. Arch. Oral Biol. 1973, 18, 957–962. [Google Scholar] [CrossRef]
- James, S.A.; Phillips, S.; Telatin, A.; Baker, D.; Ansorge, R.; Clarke, P.; Hall, L.J.; Carding, S.R. Preterm Infants Harbour a Rapidly Changing Mycobiota That Includes Candida Pathobionts. J. Fungi 2020, 6, 273. [Google Scholar] [CrossRef] [PubMed]
- Manzoni, P.; Stolfi, I.; Pugni, L.; Decembrino, L.; Magnani, C.; Vetrano, G.; Tridapalli, E.; Corona, G.; Giovannozzi, C.; Farina, D.; et al. A multicenter, randomized trial of prophylactic fluconazole in preterm neonates. N. Engl. J. Med. 2007, 356, 2483–2495. [Google Scholar] [CrossRef] [Green Version]
- Gunsalus, K.T.; Tornberg-Belanger, S.N.; Matthan, N.R.; Lichtenstein, A.H.; Kumamoto, C.A. Manipulation of Host Diet to Reduce Gastrointestinal Colonization by the Opportunistic Pathogen Candida albicans. mSphere 2016, 1, e00020-15. [Google Scholar] [CrossRef] [Green Version]
- Arsenault, A.B.; Gunsalus, K.T.W.; Laforce-Nesbitt, S.S.; Przystac, L.; DeAngelis, E.J.; Hurley, M.E.; Vorel, E.S.; Tucker, R.; Matthan, N.R.; Lichtenstein, A.H.; et al. Dietary Supplementation with Medium-Chain Triglycerides Reduces Candida Gastrointestinal Colonization in Preterm Infants. Pediatr. Infect. Dis. J. 2019, 38, 164–168. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [Green Version]
- Katoh, K.; Misawa, K.; Kuma, K.; Miyata, T. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef] [Green Version]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree 2--approximately maximum-likelihood trees for large alignments. PLoS ONE 2010, 5, e9490. [Google Scholar] [CrossRef]
- Lozupone, C.A.; Hamady, M.; Kelley, S.T.; Knight, R. Quantitative and qualitative beta diversity measures lead to different insights into factors that structure microbial communities. Appl. Environ. Microbiol. 2007, 73, 1576–1585. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McDonald, D.; Price, M.N.; Goodrich, J.; Nawrocki, E.P.; DeSantis, T.Z.; Probst, A.; Andersen, G.L.; Knight, R.; Hugenholtz, P. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 2012, 6, 610–618. [Google Scholar] [CrossRef] [PubMed]
- Friedman, J.; Alm, E.J. Inferring correlation networks from genomic survey data. PLoS Comput. Biol. 2012, 8, e1002687. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koenig, J.E.; Spor, A.; Scalfone, N.; Fricker, A.D.; Stombaugh, J.; Knight, R.; Angenent, L.T.; Ley, R.E. Succession of microbial consortia in the developing infant gut microbiome. Proc. Natl. Acad. Sci. USA 2011, 108 (Suppl. S1), 4578–4585. [Google Scholar] [CrossRef] [Green Version]
- La Rosa, P.S.; Warner, B.B.; Zhou, Y.; Weinstock, G.M.; Sodergren, E.; Hall-Moore, C.M.; Stevens, H.J.; Bennett, W.E., Jr.; Shaikh, N.; Linneman, L.A.; et al. Patterned progression of bacterial populations in the premature infant gut. Proc. Natl. Acad. Sci. USA 2014, 111, 12522–12527. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tauchi, H.; Yahagi, K.; Yamauchi, T.; Hara, T.; Yamaoka, R.; Tsukuda, N.; Watanabe, Y.; Tajima, S.; Ochi, F.; Iwata, H.; et al. Gut microbiota development of preterm infants hospitalised in intensive care units. Benef. Microbes 2019, 10, 641–651. [Google Scholar] [CrossRef]
- Schwiertz, A.; Gruhl, B.; Lobnitz, M.; Michel, P.; Radke, M.; Blaut, M. Development of the intestinal bacterial composition in hospitalized preterm infants in comparison with breast-fed, full-term infants. Pediatr. Res. 2003, 54, 393–399. [Google Scholar] [CrossRef] [Green Version]
- Arboleya, S.; Ang, L.; Margolles, A.; Yiyuan, L.; Dongya, Z.; Liang, X.; Solis, G.; Fernandez, N.; de Los Reyes-Gavilan, C.G.; Gueimonde, M. Deep 16S rRNA metagenomics and quantitative PCR analyses of the premature infant fecal microbiota. Anaerobe 2012, 18, 378–380. [Google Scholar] [CrossRef] [Green Version]
- Fehr, K.; Moossavi, S.; Sbihi, H.; Boutin, R.C.T.; Bode, L.; Robertson, B.; Yonemitsu, C.; Field, C.J.; Becker, A.B.; Mandhane, P.J.; et al. Breastmilk Feeding Practices Are Associated with the Co-Occurrence of Bacteria in Mothers’ Milk and the Infant Gut: The CHILD Cohort Study. Cell Host Microbe 2020, 28, 285–297.e4. [Google Scholar] [CrossRef]
- Azad, M.B. Infant Feeding and the Developmental Origins of Chronic Disease in the CHILD Cohort: Role of Human Milk Bioactives and Gut Microbiota. Breastfeed. Med. 2019, 14, S22–S24. [Google Scholar] [CrossRef] [Green Version]
- Pannaraj, P.S.; Li, F.; Cerini, C.; Bender, J.M.; Yang, S.; Rollie, A.; Adisetiyo, H.; Zabih, S.; Lincez, P.J.; Bittinger, K.; et al. Association Between Breast Milk Bacterial Communities and Establishment and Development of the Infant Gut Microbiome. JAMA Pediatr. 2017, 171, 647–654. [Google Scholar] [CrossRef] [PubMed]





| Control (n = 8) | MCT (n = 9) | |
|---|---|---|
| Maternal age, years * | 29 (18–36) | 27 (19–38) |
| Male gender, n (%) | 4 (50) | 6 (67) |
| Gestational age, weeks * | 28.9 (25–33) | 27.2 (23–32) |
| Cesarean delivery, n (%) | 5 (63) | 6 (67) |
| Any prenatal antibiotic, n (%) | 5 (63) | 8 (89) |
| Any prenatal steroid, n (%) | 5 (63) | 8 (89) |
| Birth weight, g * | 1229 (500–2010) | 876 (360–1220) |
| Any breast milk, n (%) | 7 (88) | 7 (78) |
| Exclusive breast milk, n (%) | 1 (13) | 3 (33) |
| Parenteral nutrition days * | 10 (4–26) | 18 (4–53) |
| Age at enrollment, days * | 27 (13–55) | 34 (9–90) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Romo, J.A.; Arsenault, A.B.; Laforce-Nesbitt, S.S.; Bliss, J.M.; Kumamoto, C.A. Minimal Effects of Medium-Chain Triglyceride Supplementation on the Intestinal Microbiome Composition of Premature Infants: A Single-Center Pilot Study. Nutrients 2022, 14, 2159. https://0-doi-org.brum.beds.ac.uk/10.3390/nu14102159
Romo JA, Arsenault AB, Laforce-Nesbitt SS, Bliss JM, Kumamoto CA. Minimal Effects of Medium-Chain Triglyceride Supplementation on the Intestinal Microbiome Composition of Premature Infants: A Single-Center Pilot Study. Nutrients. 2022; 14(10):2159. https://0-doi-org.brum.beds.ac.uk/10.3390/nu14102159
Chicago/Turabian StyleRomo, Jesús A., Amanda B. Arsenault, Sonia S. Laforce-Nesbitt, Joseph M. Bliss, and Carol A. Kumamoto. 2022. "Minimal Effects of Medium-Chain Triglyceride Supplementation on the Intestinal Microbiome Composition of Premature Infants: A Single-Center Pilot Study" Nutrients 14, no. 10: 2159. https://0-doi-org.brum.beds.ac.uk/10.3390/nu14102159