Insect Antimicrobial Peptides, a Mini Review
Abstract
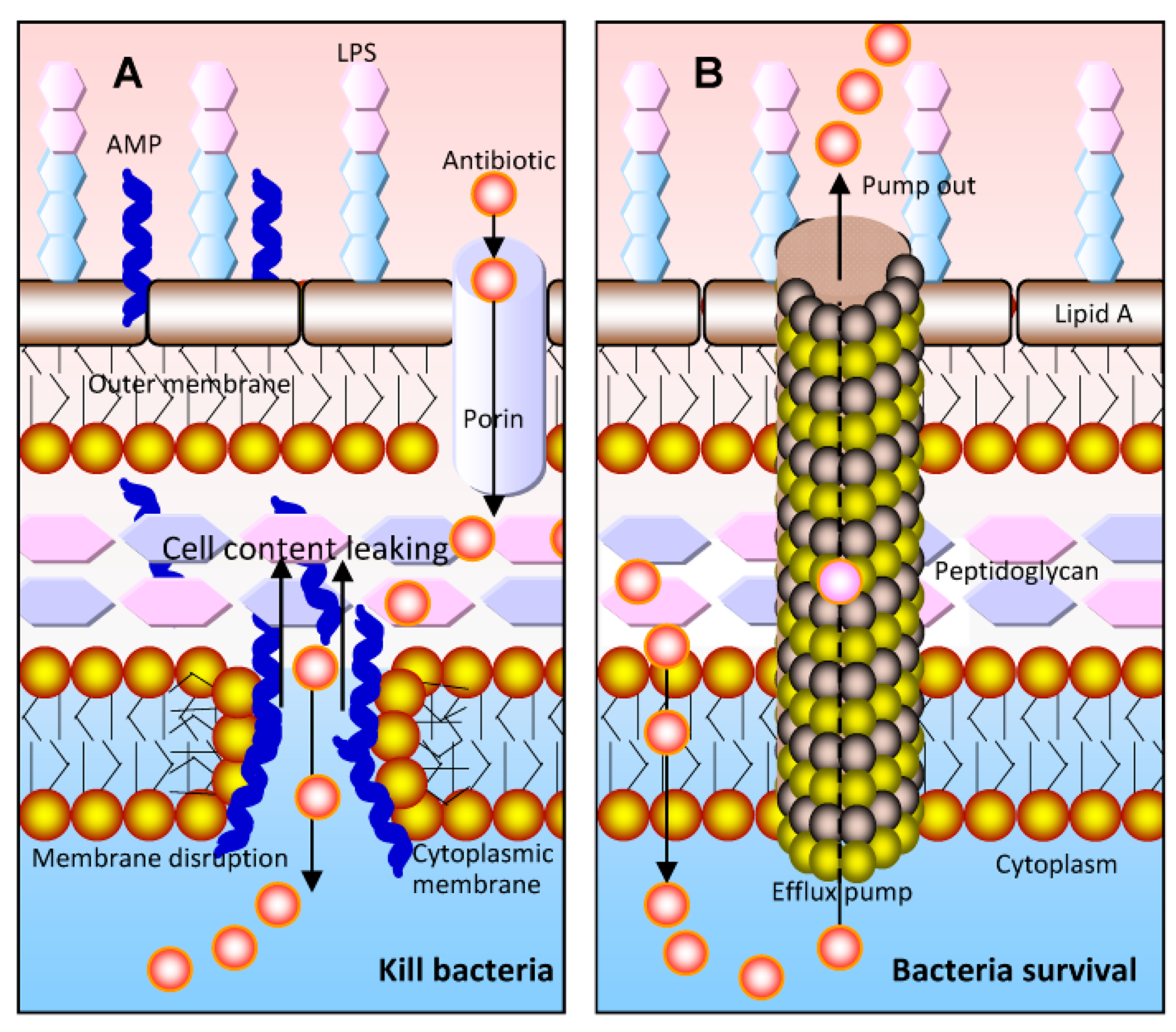
:1. Introduction
2. Insect Antimicrobial Peptides
2.1. Defensins
2.2. Cecropins
2.3. Attacins
2.4. Lebocins
2.5. Drosocin
2.6. Diptericins
2.7. Metchnikowin
2.8. Ponericins
2.9. Jelleines
2.10. Apisimin
2.11. Pyrrhocoricin
2.12. Persulcatusin
2.13. Melittin
3. Concluding Remarks
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bulet, P.; Hetru, C.; Dimarcq, J.L.; Hoffmann, D. Abtimicrobial peptides in insects; structure and function. Dev. Comp. Immun. 1999, 23, 329–344. [Google Scholar] [CrossRef]
- Yi, H.Y.; Chowdhury, M.; Huang, Y.D.; Yu, X.Q. Insect antimicrobial peptides and their applications. Appl. Microbiol. Biotechnol. 2014, 98, 5807–5822. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Uccelletti, D.; Zanni, E.; Marcellini, L.; Palleschi, C.; Barra, D.; Mangoni, M.L. Anti-Pseudomonas activity of frog skin antimicrobial peptides in a Caenorhabditis elegans infection model: A plausible mode of action in vitro and in vivo. Antimicrob. Agents Chemother. 2010, 54, 3853–3860. [Google Scholar] [CrossRef] [PubMed]
- Tonk, M.; Vilcinskas, A.; Rahnamaeian, M. Insect antimicrobial peptides: Potential tools for the prevention of skin cancer. Appl. Microbiol. Biotechnol. 2016, 100, 7397–7405. [Google Scholar] [CrossRef] [PubMed]
- Ongey, E.L.; Pflugmacher, S.; Neubauer, P. Bioinspired designs, molecular premise and tools for evaluating the ecological importance of antimicrobial peptides. Pharmaceuticals 2018, 11, 68. [Google Scholar] [CrossRef] [PubMed]
- Patocka, J.; Nepovimova, E.; Klimova, B.; Wu, Q.; Kuca, K. Antimicrobial peptides: Amphibian host defense peptides. Curr. Med. Chem. 2018. [Google Scholar] [CrossRef] [PubMed]
- Jin, G.; Weinberg, A. Human antimicrobial peptides and cancer. Semin. Cell Dev. Biol. 2018. [Google Scholar] [CrossRef] [PubMed]
- Kumar, P.; Kizhakkedathu, J.N.; Straus, S.K. Antimicrobial peptides: Diversity, mechanism of action and strategies to improve the activity and biocompatibility in vivo. Biomolecules 2018, 8, 4. [Google Scholar] [CrossRef] [PubMed]
- Chernysh, S.; Gordya, N.; Suborova, T. Insect antimicrobial peptide complexes prevent resistance development in bacteria. PLoS ONE 2015, 10, e0130788. [Google Scholar] [CrossRef] [PubMed]
- Shen, W.; He, P.; Xiao, C.; Chen, X. From Antimicrobial Peptides to Antimicrobial Poly(α-amino acid)s. Adv. Healthc. Mater. 2018, 1, 1800354. [Google Scholar] [CrossRef] [PubMed]
- Hollmann, A.; Martinez, M.; Maturana, P.; Semorile, L.C.; Maffia, P.C. Antimicrobial peptides: Interaction with model and biological membranes and synergism with chemical antibiotics. Front. Chem. 2018, 6, 204. [Google Scholar] [CrossRef] [PubMed]
- Jozefiak, A.; Engberg, R.M. Insect proteins as a potential source of antimicrobial peptides in livestock production. A review. J. Anim. Feed Sci. 2017, 26, 87–99. [Google Scholar] [CrossRef] [Green Version]
- Bechinger, B.; Gorr, S.U. Antimicrobial peptides: Mechanisms of action and resistance. J. Dent. Res. 2017, 96, 254–260. [Google Scholar] [CrossRef] [PubMed]
- Oñate-Garzón, J.; Manrique-Moreno, M.; Trier, S.; Leidy, C.; Torres, R.; Patiño, E. Antimicrobial activity and interactions of cationic peptides derived from Galleria mellonella cecropin D-like peptide with model membranes. J. Antibiot. 2017, 70, 238–245. [Google Scholar] [CrossRef] [PubMed]
- Silvestro, L.; Axelsen, P.H. Membrane-induced folding of cecropin A. Biophys. J. 2000, 79, 1465–1477. [Google Scholar] [CrossRef]
- Yeaman, M.R.; Yount, N.Y. Mechanisms of antimicrobial peptide action and resistance. Pharmacol. Rev. 2003, 55, 27–55. [Google Scholar] [CrossRef] [PubMed]
- Powers, J.P.S.; Hancock, R.E.W. The relationship between peptide structure and antibacterial activity. Peptides 2003, 24, 1681–1691. [Google Scholar] [CrossRef] [PubMed]
- Nan, Y.H.; Shin, S.Y. Effect of disulphide bond position on salt resistance and LPS-neutralizing activity of α-helical homo-dimeric model antimicrobial peptides. BMB Rep. 2011, 44, 747–752. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yaakobi, K.; Liebes-Peer, Y.; Kushmaro, A.; Rapaport, H. Designed amphiphilic β-sheet peptides as templates for paraoxon adsorption and detection. Langmuir 2013, 29, 6840–6848. [Google Scholar] [CrossRef] [PubMed]
- Freudenthal, O.; Quilès, F.; Francius, G. Discrepancies between cyclic and linear antimicrobial peptide actions on the spectrochemical and nanomechanical fingerprints of a young biofilm. ACS Omega 2017, 2, 5861–5872. [Google Scholar] [CrossRef] [PubMed]
- Kindrachuk, J.; Napper, S. Structure-activity relationships of multifunctional host defence peptides. Mini-Rev. Med. Chem. 2010, 10, 596–614. [Google Scholar] [CrossRef] [PubMed]
- Suarez, M.; Haenni, M.; Canarelli, S.; Fisch, F.; Chodanowski, P.; Servis, C.; Michielin, O.; Freitag, R.; Moreillon, P.; Mermod, N. Structure-function characterization and optimization of a plant-derived antibacterial peptide. Antimicrob. Agents Chemother. 2005, 49, 3847–3857. [Google Scholar] [CrossRef] [PubMed]
- McDonald, M.; Mannion, M.; Pike, D.; Lewis, K.; Flynn, A.; Brannan, A.M.; Browne, M.J.; Jackman, D.; Madera, L.; Power Coombs, M.R.; Hoskin, D.W.; Rise, M.L.; Booth, V. Structure-function relationships in histidine-rich antimicrobial peptides from Atlantic cod. Biochim. Biophys. Acta 2015, 1848, 1451–1461. [Google Scholar] [CrossRef] [PubMed]
- Locock, K.E.; Michl, T.D.; Valentin, J.D.; Vasilev, K.; Hayball, J.D.; Qu, Y.; Traven, A.; Griesser, H.J.; Meagher, L.; Haeussler, M. Guanylated polymethacrylates: A class of potent antimicrobial polymers with low hemolytic activity. Biomacromolecules 2013, 14, 4021–4031. [Google Scholar] [CrossRef] [PubMed]
- Leptihn, S.; Har, J.Y.; Wohland, T.; Ding, J.L. Correlation of charge, hydrophobicity, and structure with antimicrobial activity of S1 and MIRIAM peptides. Biochemistry 2010, 49, 9161–9170. [Google Scholar] [CrossRef] [PubMed]
- Naumenkova, T.V.; Antonov, M.I.u.; Nikolaev, I.N.; Shaĭtan, K.V. Effect of Pro11-Ala11 amino acid substitution on structural and functional properties of antimicrobial peptide buforin 2. Biofizika 2012, 57, 988–999. [Google Scholar] [PubMed]
- Derache, C.; Meudal, H.; Aucagne, V.; Mark, K.J.; Cadène, M.; Delmas, A.F.; Lalmanach, A.C.; Landon, C. Initial insights into structure-activity relationships of avian defensins. J. Biol. Chem. 2012, 287, 7746–7755. [Google Scholar] [CrossRef] [PubMed]
- Ahn, M.J.; Sohn, H.I.; Nan, Y.H.; Murugan, R.N.; Cheong, C.; Ryu, E.K.; Kim, E.H.; Kang, S.W.; Kim, E.J.; Shin, S.Y.; et al. Functional and structural characterization of Drosocin and its derivatives linked O-GalNAc at Thr 11 residue. Bull. Korean Chem. Soc. 2011, 32, 3327–3332. [Google Scholar] [CrossRef]
- Walkenhorst, W.F.; Klein, J.W.; Vo, P.; Wimley, W.C. pH Dependence of microbe sterilization by cationic antimicrobial peptides. Antimicrob. Agents Chemother. 2013, 57, 3312–3320. [Google Scholar] [CrossRef] [PubMed]
- Makarova, O.; Johnston, P.; Rodriguez-Rojas, A.; El Shazely, B.; Morales, J.M.; Rolff, J. Genomics of experimental adaptation of Staphylococcus aureus to a natural combination of insect antimicrobial peptides. Sci. Rep. 2018, 8, 15359. [Google Scholar] [CrossRef] [PubMed]
- Mylonakis, E.; Podsiadlowski, L.; Muhammed, M.; Vilcinskas, A. Diversity, evolution and medical applications of insect antimicrobial peptides. Philos. Trans. R. Soc. Lond. B 2016, 371, 20150290. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, L.J.; Gallo, R.L. Antimicrobial peptides. Curr. Biol. 2016, 26, 14–19. [Google Scholar] [CrossRef] [PubMed]
- Rozgonyi, F.; Szabo, D.; Kocsis, B.; Ostorházi, E.; Abbadessa, G.; Cassone, M.; Wade, J.D.; Otvos, L., Jr. The antibacterial effect of a proline-rich antibacterial peptide A3-APO. Curr. Med. Chem. 2009, 16, 3996–4002. [Google Scholar] [CrossRef] [PubMed]
- Ganz, T.; Lehrer, R.I. Defensins. Pharmacol. Ther. 1995, 66, 191–205. [Google Scholar] [CrossRef]
- Ganz, T. Defensins: Antimicrobial peptides of innate immunity. Nat. Rev. Immunol. 2003, 3, 710–720. [Google Scholar] [CrossRef] [PubMed]
- Thomma, B.P.; Cammue, B.P.; Thevissen, K. Plant defensins. Planta 2002, 216, 193–202. [Google Scholar] [CrossRef] [PubMed]
- Zhu, S.; Gao, B. Evolutionary origin of β-defensins. Dev. Comp. Immunol. 2013, 39, 79–84. [Google Scholar] [CrossRef] [PubMed]
- Cederlund, A.; Gudmundsson, G.H.; Agerberth, B. Antimicrobial peptides important in innate immunity. FEBS J. 2011, 278, 3942–3951. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tay, D.K.; Rajagopalan, G.; Li, X.; Chen, Y.; Lua, L.H.; Leong, S.S. A new bioproduction route for a novel antimicrobial peptide. Biotechnol. Bioeng. 2011, 108, 572–581. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, J.A.; Hetru, C. Insect defensins: Inducible antibacterial peptides. Immunol. Today 1992, 13, 411–415. [Google Scholar] [CrossRef]
- Hetru, C.; Troxler, L.; Hoffmann, J.A. Drosophiola melanogaster antimicrobial defense. J. Infect. Dis. 2003, 187, S327–S343. [Google Scholar] [CrossRef] [PubMed]
- Gomes, P.S.; Fernandes, M.H. Defensins in the oral cavity: Distribution and biological role. J. Oral Pathol. Med. 2010, 39, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Boman, H.G. Antibacterial peptides: Key components needed in immunity. Cell 1991, 65, 205–207. [Google Scholar] [CrossRef]
- Zhao, W.Y.; Dong, B.R.; Zhou, Y. In vitro antimicrobial activity of defensins from rabbit neutrophils against Pseudomonas aeruginosa and its multiple-drug-resistance strains. Sichuan Da Xue Xue Bao Yi Xue Ban. 2005, 36, 83–85. [Google Scholar] [PubMed]
- Fujiwara, S.; Imai, J.; Fujiwara, M.; Yaeshima, T.; Kawashima, T.; Kobayashi, K. A potent antibacterial protein in royal jelly. J. Biol. Chem. 1990, 265, 11333–11337. [Google Scholar] [PubMed]
- Bılikova, K.; Gusu, W.; Simuth, J. Isolation of a peptide fraction from honeybee royal jelly as a potential antifoulbrood factor. Apidologie 2001, 32, 275–283. [Google Scholar] [CrossRef]
- Steiner, H.; Hultmark, D.; Engstrom, A.; Bennich, H.; Boman, H.G. Sequence and specificity of two antibacterial proteins involved in insect immunity. Nature 1981, 292, 246–248. [Google Scholar] [CrossRef] [PubMed]
- Hultmark, D.; Engström, A.; Bennich, H.; Kapur, R.; Boman, H.G. Insect immunity. Isolation and structure of cecropin D and four minor antibacterial components from cecropia pupae. Eur. J. Biochem. 1982, 127, 207–217. [Google Scholar] [CrossRef] [PubMed]
- Van Hofsten, P.; Faye, I.; Kockum, K.; Lee, J.Y.; Xanthopoulos, K.G.; Boman, I.A. Molecular cloning, cDNA sequencing, and chemical synthesis of cecropin B from Hyalophora cecropia. Proc. Nat. Acad. Sci. USA 1985, 82, 2240–2243. [Google Scholar] [CrossRef] [PubMed]
- Moore, A.J.; Beazley, W.D.; Bibby, M.C.; Devine, D.A. Antimicrobial activity of cecropins. J. Antimicrobiol. Chemother. 1996, 37, 1077–1089. [Google Scholar] [CrossRef] [Green Version]
- Bechinger, B.; Lohner, K. Detergent-like actions of linear amphipathic cationic antimicrobial peptides. Biochim. Biophys. Acta 2006, 1758, 1529–1539. [Google Scholar] [CrossRef] [PubMed]
- Ouyang, L.; Xu, X.; Freed, S.; Gao, Y.; Yu, J.; Wang, S.; Ju, W.; Zhang, Y.; Jin, F. Cecropins from Plutella xylostella and Their Interaction with Metarhizium anisopliae. PLoS ONE 2015, 10, e0142451. [Google Scholar] [CrossRef] [PubMed]
- Srisailam, S.; Kumar, T.K.S.; Arunkumar, A.I.; Leung, K.W.; Yu, C.; Chen, H.M. Crumpled structure of the custom hydrophobic lytic peptide cecropin B3. Eur. J. Biochem. 2001, 268, 4278–4284. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pillai, A.; Ueno, S.; Zhang, H.; Lee, J.M.; Kato, Y. Cecropin P1 and novel nematode cecropins: A bacteria-inducible antimicrobial peptide family in the nematode Ascaris suum. Biochem. J. 2005, 390, 207–214. [Google Scholar] [CrossRef] [PubMed]
- Fu, H.; Björstad, A.; Dahlgren, C.; Bylund, J. A bacterial cecropin-A peptide with a stabilized α-helical structure possess an increased killing capacity but no proinflammatory activity. Inflammation 2004, 28, 337–343. [Google Scholar] [CrossRef] [PubMed]
- Yun, J.; Lee, D.G. Cecropin A-induced apoptosis is regulated by ion balance and glutathione antioxidant system in Candida albicans. IUBMB Life 2016, 68, 652–662. [Google Scholar] [CrossRef] [PubMed]
- Durell, S.R.; Raghunathan, G.; Guy, H.R. Modeling the ion channel structure of cecropin. Biophys. J. 1992, 63, 1623–1631. [Google Scholar] [CrossRef] [Green Version]
- Lu, D.; Geng, T.; Hou, C.; Huang, Y.; Qin, G.; Guo, X. Bombyx mori cecropin A has a high antifungal activity to entomopathogenic fungus Beauveria bassiana. Gene 2016, 583, 29–35. [Google Scholar] [CrossRef] [PubMed]
- Srisailam, S.; Arunkumar, A.I.; Wang, W.; Yu, C.; Chen, H.M. Conformational study of a custom antibacterial peptide cecropin B1: Implications of the lytic activity. Biochim. Biophys. Acta 2000, 1479, 275–285. [Google Scholar] [CrossRef]
- Giacometti, A.; Cirioni, O.; Ghiselli, R.; Viticchi, C.; Mocchegiani, F.; Riva, A.; Saba, V.; Scalise, G. Effect of mono-dose intraperitoneal cecropins in experimental septic shock. Crit. Care Med. 2001, 29, 1666–1669. [Google Scholar] [CrossRef] [PubMed]
- Chalk, R.; Townson, H.; Ham, P.J. Brugia pahangi: The effects of cecropins on microfilariae in vitro and in Aedes aegypti. Exp. Parasitol. 1995, 80, 401–406. [Google Scholar] [CrossRef] [PubMed]
- Andrä, J.; Berninghausen, O.; Leippe, M. Cecropins, antibacterial peptides from insects and mammals, are potently fungicidal against Candida albicans. Med. Microbiol. Immunol. 2001, 189, 169–173. [Google Scholar] [CrossRef] [PubMed]
- Hara, S.; Yamakawa, M. A novel antibacterial peptide family isolated from the silkworm, Bombyx mori. Biochem. J. 1995, 310, 651–656. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hara, S.; Yamakawa, M. Moricin, a novel type of antibacterial peptide isolated from the silkworm, Bombyx mori. J. Biol. Chem. 1995, 270, 29923–29927. [Google Scholar] [PubMed]
- Hu, H.; Wang, C.; Guo, X.; Li, W.; Wang, Y.; He, Q. Broad activity against porcine bacterial pathogens displayed by two insect antimicrobial peptides moricin and cecropin B. Mol. Cells 2013, 35, 106–114. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gudmundsson, G.H.; Lidholm, D.A.; Asling, B.; Gan, R.; Boman, H.G. The cecropin locus. Cloning and expression of a gene cluster encoding three antibacterial peptides in Hyalophora cecropia. J. Biol. Chem. 1991, 266, 11510–11517. [Google Scholar] [PubMed]
- Guo, C.; Huang, Y.; Zheng, H.; Tang, L.; He, J.; Xiao, L.; Liu, D.; Jiang, H. Secretion and activity of antimicrobial peptide cecropin D expressed in Pichia pastoris. Exp. Ther. Med. 2012, 4, 1063–1068. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, S.I.; An, H.S.; Chang, B.S.; Yoe, S.M. Expression, cDNA cloning, and characterization of the antibacterial peptide cecropin D from Agrius convolvuli. Anim. Cells Syst. 2013, 17, 23–30. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.; Guo, C.; Huang, Y.; Zhang, X.; Chen, Y. Inhibition of porcine reproductive and respiratory syndrome virus by Cecropin D in vitro. Infect. Genet. Evol. 2015, 34, 7–16. [Google Scholar] [CrossRef] [PubMed]
- Hou, Z.P.; Wang, W.J.; Liu, Z.Q.; Liu, G.; Souffrant, W.B.; Yin, Y.L. Effect of lactoferricin B and cecropin P1 against enterotoxigenic Escherichia coli in vitro. J. Food Agric. Environ. 2011, 9, 271–274. [Google Scholar]
- Baek, M.H.; Kamiya, M.; Kushibiki, T.; Nakazumi, T.; Tomisawa, S.; Abe, C.; Kumaki, Y.; Kikukawa, T.; Demura, M.; Kawano, K.; et al. Lipopolysaccharide-bound structure of the antimicrobial peptide cecropin P1 determined by nuclear magnetic resonance spectroscopy. J. Pept. Sci. 2016, 22, 214–221. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arcidiacono, S.; Soares, J.W.; Meehan, A.M.; Marek, P.; Kirby, R. Membrane permeability and antimicrobial kinetics of cecropin P1 against Escherichia coli. J. Pept. Sci. 2009, 15, 398–403. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Wei, P.H.; Zhu, X.; Wirth, M.J.; Bhunia, A.; Narsimhan, G. Effect of immobilization on the antimicrobial activity of a cysteine-terminated antimicrobial Peptide Cecropin P1 tethered to silica nanoparticle against E. coli O157:H7 E.D.L. Colloids Surf. B Biointerfaces 2017, 156, 305–312. [Google Scholar] [CrossRef] [PubMed]
- Zakharchenko, N.S.; Belous, A.S.; Biryukova, Y.K.; Medvedeva, OA.; Belyakova, A.V.; Masgutova, G.A.; Trubnikova, E.V.; Buryanov, Y.I.; Lebedeva, A.A. Immunomodulating and Revascularizing Activity of Kalanchoe pinnata Synergize with Fungicide Activity of Biogenic Peptide Cecropin P1. J. Immunol. Res. 2017, 2017, 3940743. [Google Scholar] [CrossRef] [PubMed]
- Guo, C.; Huang, Y.; Cong, P.; Liu, X.; Chen, Y.; He, Z. Cecropin P1 inhibits porcine reproductive and respiratory syndrome virus by blocking attachment. BMC Microbiol. 2014, 14, 273. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Han, X.; He, N.; Chen, Z.; Brooks, C.L., 3rd. Molecular structures of C- and N-terminus cysteine modified cecropin P1 chemically immobilized onto maleimide-terminated self-assembled monolayers investigated by molecular dynamics simulation. J. Phys. Chem. B 2014, 118, 5670–5680. [Google Scholar] [CrossRef] [PubMed]
- Téllez, G.A.; Castaño-Osorio, J.C. Expression and purification of an active cecropin-like recombinant protein against multidrug resistance Escherichia coli. Protein Expr. Purif. 2014, 100, 48–53. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; Shen, J.; Jin, X.; Ma, Y.; Huang, Y.; Mei, H.; Chu, F.; Zhu, J. Bactericidal activity of Musca domestica cecropin (Mdc) on multidrug-resistant clinical isolate of Escherichia coli. Appl. Microbiol. Biotechnol. 2012, 95, 939–945. [Google Scholar] [CrossRef] [PubMed]
- Hultmark, D.; Engström, A.; Andersson, K.; Steiner, H.; Bennich, H.; Boman, H.G. Insect immunity. Attacins, a family of antibacterial proteins from Hyalophora cecropia. EMBO J. 1983, 2, 571–576. [Google Scholar] [CrossRef] [PubMed]
- Carlsson, A.; Engström, P.; Palva, E.T.; Bennich, H. Attacin, an antibacterial protein from Hyalophora cecropia, inhibits synthesis of outer membrane proteins in Escherichia coli by interfering with omp gene transcription. Infect. Immun. 1991, 59, 3040–3045. [Google Scholar] [PubMed]
- Carlsson, A.; Nyström, T.; de Cock, H.; Bennich, H. Attacin—An insect immune protein—Binds LPS and triggers the specific inhibition of bacterial outer-membrane protein synthesis. Microbiology 1998, 144, 2179–2188. [Google Scholar] [CrossRef] [PubMed]
- Kockum, K.; Faye, I.; Hofsten, P.V.; Lee, J.Y.; Xanthopoulos, K.G.; Boman, H.G. Insect immunity. Isolation and sequence of two cDNA clones corresponding to acidic and basic attacins from Hyalophora cecropia. EMBO J. 1984, 3, 2071–2075. [Google Scholar] [CrossRef] [PubMed]
- Dushay, M.S.; Roethele, J.B.; Chaverri, J.M.; Dulek, D.E.; Syed, S.K.; Kitami, T.; Eldon, E.D. Two attacin antibacterial genes of Drosophila melanogaster. Gene 2000, 246, 49–57. [Google Scholar] [CrossRef]
- Geng, H.; An, C.J.; Hao, Y.J.; Li, D.S.; Du, R.Q. Molecular cloning and expression of Attacin from housefly (Musca domestica). Yi Chuan Xue Bao 2004, 31, 1344–1350. [Google Scholar] [PubMed]
- Liu, G.; Kang, D.; Steiner, H. Trichoplusia ni lebocin, an inducible immune gene with a downstream insertion element. Biochem. Biophys. Res. Commun. 2000, 269, 803–807. [Google Scholar] [CrossRef] [PubMed]
- Casteels, P.; Ampe, C.; Riviere, L.; Van Damme, J.; Elicone, C.; Fleming, M.; Jacobs, F.; Tempst, P. Isolation and characterization of abaecin, a major antibacterial response peptide in the honeybee (Apis mellifera). Eur. J. Biochem. 1990, 187, 381–386. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Slocinska, M.; Marciniak, P.; Rosinski, G. Insects antiviral and anticancer peptides: New leads for the future? Protein Pept. Lett. 2008, 15, 578–585. [Google Scholar] [CrossRef] [PubMed]
- Bulet, P.; Dimarcq, J.L.; Hetru, C.; Lagueux, M.; Charlet, M.; Hegy, G.; Van Dorsselaer, A.; Hoffmann, J.A. A novel inducible antibacterial peptide of Drosophila carries an O-glycosylated substitution. J. Biol. Chem. 1993, 268, 14893–14897. [Google Scholar] [PubMed]
- McManus, A.M.; Otvos, L., Jr.; Hoffmann, R.; Craik, D.J. Conformational studies by NMR of the antimicrobial peptide, drosocin, and its non-glycosylated derivative: Effects of glycosylation on solution conformation. Biochemistry 1999, 38, 705–714. [Google Scholar] [CrossRef] [PubMed]
- Lele, D.S.; Talat, S.; Kumari, S.; Srivastava, N.; Kaur, K.J. Understanding the importance of glycosylated threonine and stereospecific action of Drosocin, a Proline rich antimicrobial peptide. Eur. J. Med. Chem. 2015, 92, 637–647. [Google Scholar] [CrossRef] [PubMed]
- Bulet, P.; Urge, L.; Ohresser, S.; Hetru, C.; Otvos, L., Jr. Enlarged scale chemical synthesis and range of activity of drosocin, an O-glycosylated antibacterial peptide of Drosophila. Eur. J. Biochem. 1996, 238, 64–69. [Google Scholar] [CrossRef] [PubMed]
- Imler, J.L.; Bulet, P. Antimicrobial peptides in Drosophila: Structures, activities and gene regulation. Chem. Immunol. Allergy 2005, 86, 1–21. [Google Scholar] [PubMed]
- Gobbo, M.; Biondi, L.; Filira, F.; Gennaro, R.; Benincasa, M.; Scolaro, B.; Rocchi, R. Antimicrobial peptides: Synthesis and antibacterial activity of linear and cyclic drosocin and apidaecin 1b analogues. J. Med. Chem. 2002, 45, 4494–4504. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, K.; Orikasa, Y.; Ichinohe, K.; Hashimoto, S.; Ooi, T.; Taguchi, S. Flow cytometric analysis of the contributing factors for antimicrobial activity enhancement of cell-penetrating type peptides: Case study on engineered apidaecins. Biochem. Biophys. Res. Commun. 2010, 395, 7–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bluhm, M.E.; Schneider, V.A.; Schäfer, I.; Piantavigna, S.; Goldbach, T.; Knappe, D.; Seibel, P.; Martin, L.L.; Veldhuizen, E.J.; Hoffmann, R. N-Terminal Ile-Orn- and Trp-Orn-Motif Repeats Enhance Membrane Interaction and Increase the Antimicrobial Activity of Apidaecins against Pseudomonas aeruginosa. Front. Cell Dev. Biol. 2016, 4, 39. [Google Scholar] [CrossRef] [PubMed]
- Cudic, M.; Bulet, P.; Hoffmann, R.; Craik, D.J.; Otvos, L., Jr. Chemical synthesis, antibacterial activity and conformation of diptericin, an 82-mer peptide originally isolated from insects. Eur. J. Biochem. 1999, 266, 549–558. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dimarcq, J.L.; Keppi, E.; Dunbar, B.; Lambert, J.; Reichhart, J.M.; Hoffmann, D.; Rankine, S.M.; Fothergill, J.E.; Hoffmann, J.A. Insect immunity. Purification and characterization of a family of novel inducible antibacterial proteins from immunized larvae of the dipteran Phormia terranovae and complete amino-acid sequence of the predominant member, diptericin A. Eur. J. Biochem. 1988, 171, 17–22. [Google Scholar] [CrossRef] [PubMed]
- Reichhart, J.M.; Meister, M.; Dimarcq, J.L.; Zachary, D.; Hoffmann, D.; Ruiz, C.; Richards, G.; Hoffmann, J.A. Insect immunity: Developmental and inducible activity of the Drosophila diptericin promoter. EMBO J. 1992, 11, 1469–1477. [Google Scholar] [CrossRef] [PubMed]
- Ishikawa, M.; Kubo, T.; Natori, S. Purification and characterization of a diptericin homologue from Sarcophaga peregrina (flesh fly). Biochem. J. 1992, 287, 573–578. [Google Scholar] [CrossRef] [PubMed]
- Keppi, E.; Pugsley, A.P.; Lambert, J.; Wicker, C.; Dimarcq, J.L.; Hoffmann, J.A.; Hoffmann, D. Mode of action of diptericin A, a bactericidal peptide induced in the hemolymph of Phormia terranovae larvae. Insect Biochem. Physiol. 1989, 10, 229–239. [Google Scholar] [CrossRef]
- Ursic-Bedoya, R.; Buchhop, J.; Joy, J.B.; Durvasula, R.; Lowenberger, C. Prolixicin: A novel antimicrobial peptide isolated from Rhodnius prolixus with differential activity against bacteria and Trypanosoma cruzi. Insect Mol. Biol. 2011, 20, 775–786. [Google Scholar] [CrossRef] [PubMed]
- Levashina, E.A.; Ohresser, S.; Bulet, P.; Reichhart, J.M.; Hetru, C.; Hoffmann, J.A. Metchnikowin, a novel immune-inducible proline-rich peptide from Drosophila with antibacterial and antifungal properties. Eur. J. Biochem. 1995, 233, 694–700. [Google Scholar] [CrossRef] [PubMed]
- Levashina, E.A.; Ohresser, S.; Lemaitre, B.; Imler, J.L. Two distinct pathways can control expression of the gene encoding the Drosophila antimicrobial peptide metchnikowin. J. Mol. Biol. 1998, 278, 515–527. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moghaddam, M.R.B.; Gross, T.; Becker, A.; Vilcinskas, A.; Rahnamaeian, M. The selective antifungal activity of Drosophila melanogaster metchnikowin reflects the speciesdependent inhibition of succinate-coenzyme Q reductase. Sci. Rep. 2017, 7, 8192. [Google Scholar] [CrossRef] [PubMed]
- Orivel, J.; Redeker, V.; Le Caer, J.P.; Krier, F.; Revol-Junelles, A.M.; Longeon, A.; Chaffotte, A.; Dejean, A.; Rossier, J. Ponericins, new antibacterial and insecticidal peptides from the venom of the ant Pachycondyla goeldii. J. Biol. Chem. 2001, 276, 17823–17829. [Google Scholar] [CrossRef] [PubMed]
- Johnson, S.R.; Copello, J.A.; Evans, M.S.; Suarez, A.V. A biochemical characterization of the major peptides from the Venom of the giant Neotropical hunting ant Dinoponera australis. Toxicon 2010, 55, 702–710. [Google Scholar] [CrossRef] [PubMed]
- Romanelli, A.; Moggio, L.; Montella, R.C.; Campiglia, P.; Iannaccone, M.; Capuano, F.; Pedone, C.; Capparelli, R. Peptides from Royal Jelly: Studies on the antimicrobial activity of jelleins, jelleins analogs and synergy with temporins. J. Pept. Sci. 2011, 17, 348–352. [Google Scholar] [CrossRef] [PubMed]
- Fontana, R.; Mendes, M.A.; de Souza, B.M.; Konno, K.; César, L.M.; Malaspina, O.; Palma, M.S. elleines: A family of antimicrobial peptides from the Royal Jelly of honeybees (Apis mellifera). Peptides 2004, 5, 919–928. [Google Scholar] [CrossRef] [PubMed]
- Jia, F.; Wang, J.; Peng, J; Zhao, P.; Kong, Z.; Wang, K.; Yan, W.; Wang, R. The in vitro, in vivo antifungal activity and the action mode of Jelleine-I against Candida species. Amino Acids 2018, 50, 229–239. [Google Scholar] [CrossRef] [PubMed]
- Barnutiu, L.I.; Marghitas, L.A.; Dezmirean, D.S.; Bobis, O.; Mihai, C.M.; Pavel, C. Antimicrobial compounds of Royal Jelly. Bull. Univ. Agric. Sci. Vet. Med. Cluj-Napoca 2011, 68, 85–90. [Google Scholar]
- Bilikova, K.; Hanes, J.; Nordhoff, E.; Saenger, W.; Klaudiny, J.; Simuth, J. Apisimin. A new serine-valine-rich peptide from honeybee (Apis mellifera L.) royal jelly: Purification and molecular characterization. FEBS Lett. 2002, 528, 125–129. [Google Scholar] [CrossRef]
- Shen, L.L.; Xing, Y.Y.; Gao, Q. Sequence analysis of functional Apisimin-2 cDNA from royal jelly of Chinese honeybee and its expression in Escherichia coli. Asia Pac. J. Clin. Nutr. 2007, 16, 222–226. [Google Scholar] [PubMed]
- Gannabathula, S.; Krissansen, G.W.; Skinner, M.; Steinhorn, G.; Schlothauer, R. Honeybee apisimin and plant arabinogalactans in honey costimulate monocytes. Food Chem. 2015, 168, 34–40. [Google Scholar] [CrossRef] [PubMed]
- Cociancich, S.; Dupont, A.; Hegy, G.; Lanot, R.; Holder, F.; Hetru, C.; Hoffmann, J.A.; Bulet, P. Novel inducible antibacterial peptides from a hemipteran insect, the sap-sucking bug Pyrrhocoris apterus. Biochem. J. 1994, 300, 567–575. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kragol, G.; Lovas, S.; Varadi, G.; Condie, B.A.; Hoffmann, R.; Otvos, L., Jr. The antibacterial peptide pyrrhocoricin inhibits the ATPase actions of DnaK and prevents chaperone-assisted protein folding. Biochemistry 2001, 40, 3016–3026. [Google Scholar] [CrossRef]
- Chesnokova, L.S.; Slepenkov, S.V.; Witt, S.N. The insect antimicrobial peptide, L-pyrrhocoricin, binds to and stimulates the ATPase activity of both wild-type and lidless DnaK. FEBS Lett. 2004, 565, 65–69. [Google Scholar] [CrossRef] [PubMed]
- Kragol, G.; Hoffmann, R.; Chattergoon, M.A.; Lovas, S.; Cudic, M.; Bulet, P.; Condie, B.A.; Rosengren, K.J.; Montaner, L.J.; Otvos, L., Jr. Identification of crucial residues for the antibacterial activity of the proline-rich peptide, pyrrhocoricin. Eur. J. Biochem. 2002, 269, 4226–4237. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boxell, A.; Lee, S.H.; Jefferies, R.; Watt, P.; Hopkins, R.; Reid, S.; Armson, A.; Ryan, U. Pyrrhocoricin as a potential drug delivery vehicle for Cryptosporidium parvum. Exp. Parasitol. 2008, 119, 301–303. [Google Scholar] [CrossRef] [PubMed]
- Rosengren, K.J.; Göransson, U.; Otvos, L., Jr.; Craik, D.J. Cyclization of pyrrhocoricin retains structural elements crucial for the antimicrobial activity of the native peptide. Biopolymers 2004, 76, 446–458. [Google Scholar] [CrossRef] [PubMed]
- Saito, Y.; Konnai, S.; Yamada, S.; Imamura, S.; Nishikado, H.; Ito, T.; Onuma, M.; Ohashi, K. Identification and characterization of antimicrobial peptide, defensin, in the taiga tick, Ixodes persulcatus. Insect Mol. Biol. 2009, 18, 531–539. [Google Scholar] [CrossRef] [PubMed]
- Rudenko, N.; Golovchenko, M.; Grubhoffer, L. Gene organization of a novel defensin of Ixodes ricinus: First annotation of an intron/exon structure in a hard tick defensin gene and first evidence of the occurrence of two isoforms of one member of the arthropod defensin family. Insect Mol. Biol. 2007, 16, 501–507. [Google Scholar] [CrossRef] [PubMed]
- Tsuji, N.; Battsetseg, B.; Boldbaatar, D.; Miyoshi, T.; Xuan, X.; Oliver, J.H., Jr.; Fujisaki, K. Babesial vector tick defensin against Babesia sp. parasites. Infect. Immun. 2007, 75, 3633–3640. [Google Scholar] [CrossRef] [PubMed]
- Miyoshi, N.; Isogai, E.; Hiramatsu, K.; Sasaki, T. Activity of tick antimicrobial peptide from Ixodes persulcatus (persulcatusin) against cell membranes of drug-resistant Staphylococcus aureus. J. Antibiot. 2017, 70, 142–146. [Google Scholar] [CrossRef] [PubMed]
- Miyoshi, N.; Saito, T.; Ohmura, T.; Kuroda, K.; Suita, K.; Ihara, K.; Isogai, E. Functional structure and antimicrobial activity of persulcatusin, an antimicrobial peptide from the hard tick Ixodes persulcatus. Parasites Vectors 2016, 9, 85. [Google Scholar] [CrossRef] [PubMed]
- Jamasbi, E.; Lucky, S.S.; Li, W.; Hossain, M.A.; Gopalakrishnakone, P.; Separovic, F. Effect of dimerized melittin on gastric cancer cells and antibacterial activ ity. Amino Acids 2018. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Guan, S.M.; Sun, W.; Fu, H. Melittin, the major pain-producing substance of bee venom. Neurosci. Bull. 2016, 32, 265–272. [Google Scholar] [CrossRef] [PubMed]
- Lee, G.; Bae, H. Anti-Inflammatory applications of melittin, a major component of bee venom: Detailed mechanism of action and adverse effects. Molecules 2016, 21, E616. [Google Scholar] [CrossRef] [PubMed]
- Socarras, K.M.; Theophilus, P.A.S.; Torres, J.P.; Gupta, K.; Sapi, E. Antimicrobial activity of bee venom and melittin against borrelia burgdorferi. Antibiotics 2017, 6, 31. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Singh, A.K.; Wu, X.; Lyu, Y.; Bhunia, A.K.; Narsimhan, G. Characterization of antimicrobial activity against Listeria and cytotoxicity of native melittin and its mutant variants. Colloids. Surf. B Biointerfaces 2016, 143, 194–205. [Google Scholar] [CrossRef] [PubMed]
- Leandro, L.F.; Mendes, C.A.; Casemiro, L.A.; Vinholis, A.H.; Cunha, W.R.; de Almeida, R.; Martins, C.H. Antimicrobial activity of apitoxin, melittin and phospholipase A₂ of honey bee (Apis mellifera) venom against oral pathogens. Anais Acad. Bras. Ciênc. 2015, 87, 147–155. [Google Scholar] [CrossRef] [PubMed]
- Picoli, T.; Peter, C.M.; Zani, J.L.; Waller, S.B.; Lopes, M.G.; Boesche, K.N.; Vargas, G.D.Á.; Hübner, S.O.; Fischer, G. Melittin and its potential in the destruction and inhibition of the biofilm formation by Staphylococcus aureus, Escherichia coli and Pseudomonas aeruginosa isolated from bovine milk. Microb. Pathog. 2017, 112, 57–62. [Google Scholar] [CrossRef] [PubMed]
- Shi, W.; Li, C.; Li, M.; Zong, X.; Han, D.; Chen, Y. Antimicrobial peptide melittin against Xanthomonas oryzae pv. oryzae, the bacterial leaf blight pathogen in rice. Appl. Microbiol. Biotechnol. 2016, 100, 5059–5067. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, S.K.; Ma, Q.; Li, S.B.; Gao, H.W.; Tan, Y.X.; Gong, F.; Ji, S.P. RV-23, a melittin-related peptide with cell-selective antibacterial activity and high hemocompatibility. J. Microbiol. Biotechnol. 2016, 26, 1046–1056. [Google Scholar] [CrossRef] [PubMed]
- Wiedman, G.; Fuselier, T.; He, J.; Searson, P.C.; Hristova, K.; Wimley, W.C. Highly efficient macromolecule-sized poration of lipid bilayers by a synthetically evolved peptide. J. Am. Chem. Soc. 2014, 136, 4724–4731. [Google Scholar] [CrossRef] [PubMed]
- Rady, I.; Siddiqui, I.A.; Rady, M.; Mukhtar, H. Melittin, a major peptide component of bee venom, and its conjugates in cancer therapy. Cancer Lett. 2017, 402, 16–31. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jamasbi, E.; Mularski, A.; Separovic, F. Model membrane and cell studies of antimicrobial activity of melittin analogues. Curr. Top. Med. Chem. 2016, 16, 40–45. [Google Scholar] [CrossRef] [PubMed]
- Wimley, W.C. How does melittin permeabilize membranes? Biophys. J. 2018, 114, 251–253. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.T.; Sun, T.L.; Hung, W.C.; Huang, H.W. Process of inducing pores in membranes by melittin. Proc. Natl. Acad. Sci. USA 2013, 110, 14243–14248. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wiedman, G.; Herman, K.; Searson, P.; Wimley, W.C.; Hristova, K. The electrical response of bilayers to the bee venom toxin melittin: Evidence for transient bilayer permeabilization. Biochim. Biophys. Acta-Biomembr. 2013, 1828, 1357–1364. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, J.; Lee, D.G. Melittin triggers apoptosis in Candida albicans through the reactive oxygen species-mediated mitochondria/caspasedependent pathway. FEMS Microbiol. Lett. 2014, 355, 36–42. [Google Scholar] [CrossRef] [PubMed]
- Akbari, R.; Hakemi Vala, M.; Pashaie, F.; Bevalian, P.; Hashemi, A.; Pooshang Bagheri, K. Highly synergistic effects of melittin with conventional antibiotics against multidrug-resistant isolates of acinetobacter baumannii and pseudomonas aeruginosa. Microb. Drug Resist. 2018. [Google Scholar] [CrossRef] [PubMed]
- Akbari, R.; Hakemi Vala, M.; Hashemi, A.; Aghazadeh, H.; Sabatier, J.M.; Pooshang Bagheri, K. Action mechanism of melittin-derived antimicrobial peptides, MDP1 and MDP2, de novo designed against multidrug resistant bacteria. Amino Acids 2018, 50, 1231–1243. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.H.; Jang, A.Y.; Lin, S.; Lim, S.; Kim, D.; Park, K.; Han, S.M.; Yeo, J.H.; Seo, H.S. Melittin, a honeybee venom-derived antimicrobial peptide, may target methicillin-resistant Staphylococcus aureus. Mol. Med. Rep. 2015, 12, 6483–6490. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moreno, M.; Giralt, E. Three valuable peptides from bee and wasp venoms for therapeutic and biotechnological use: Melittin, apamin and mastoparan. Toxins 2015, 7, 1126–1150. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Hou, C.; Shamsi, I.H.; Ali, E.; Muhammad, N.; Shah, J.M.; Abid, A.A. Identification of super antibiotic-resistant bacteria in diverse soils. Int. J. Agric. Biol. 2015, 17, 1133–1140. [Google Scholar] [CrossRef]
- Lee, J.K.; Luchian, T.; Park, Y. New antimicrobial peptide kills drug-resistant pathogens without detectable resistance. Oncotarget 2018, 9, 15616–15634. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Li, Z.; Li, X.; Tian, Y.; Fan, Y.; Yu, C.; Zhou, B.; Liu, Y.; Xiang, R.; Yang, L. Synergistic effects of antimicrobial peptide DP7 combined with antibiotics against multidrug-resistant bacteria. Drug Des. Devel. Ther. 2017, 11, 939–946. [Google Scholar] [CrossRef] [PubMed]
- Zerweck, J.; Strandberg, E.; Burck, J. Reichert, J.; Wadhwani, P.; Kukharenko, O.; Ulrich, A.S. Homo- and heteromeric interaction strengths of the synergistic antimicrobial peptides PGLa and magainin 2 in membranes. Eur. Biophys. J. Biophy. Lett. 2016, 45, 535–547. [Google Scholar] [CrossRef] [PubMed]
- Biswaro, L.S.; da Costa Sousa, M.G.; Rezende, T.M.B.; Dias, S.C.; Franco, O.L. Antimicrobial peptides and nanotechnology, recent advances and challenges. Front. Microbiol. 2018, 9, 855. [Google Scholar] [CrossRef] [PubMed]

| Name | Amino Acid Sequence | Reference |
|---|---|---|
| Cecropin A | GGLKKLGKKLEGVGKRVFKASEKALPVAVGIKALG-NH2 | [47] |
| Cecropin B | KWKVFKKIEKMGRNIRNGIVKAGPAIAVLGEAKAL-NH2 | [47] |
| Cecropin B1 | KWKVFKKIEKMGRNIRNGIVKAGPKWKVFKKIEK-NH2 | [53] |
| Cecropin B3 | AIAVLGEAKALMGRNIRNGIVKAGPAIAVLGEAKAL-NH2 | [53] |
| Cecropin C | GWLKKLGKRIERIGQHTRDATIQGLGIAQQAANVAATAR-NH2 | [48] |
| Cecropin D | WNPFKELEKVGQRVRDAVISAGPAVATVAQATALAK-NH2 | [48] |
| Cecropin P1 | SWLSKTAKKLENSAKKRISEGIAIAIQGGPR-NH2 | [54] |
| Name | Amino Acid Sequence |
|---|---|
| Ponericin G1 | GWKDWAKKAGGWLKKKGPGMAKAALKAAMQ-NH2 |
| Ponericin G2 | GWKDWLKKGKEWLKAKGPGIVKAALQAATQ-NH2 |
| Ponericin G3 | GWKDWLNKGKEWLKKKGPGIMKAALKAATQ-NH2 |
| Ponericin G4 | DFKDWMKTAGEWLKKKGPGILKAAMAAAT-NH2 |
| Ponericin G5 | GLKDWVKIAGGWLKKGPGILKAAMAAATQ-NH2 |
| Ponericin G6 | GLVDVLGKVGGLIKKLLP-NH2 |
| Ponericin G7 | GLVDVLGKVG GLIKKLLPG-NH2 |
| Ponericin W1 | WLGSALKIGAKLLPSVVGLFKKKKQ-NH2 |
| Ponericin W2 | WLGSALKIGAKLLPSVVGLFQKKKK-NH2 |
| Ponericin W3 | GIWGTLAKIGIKAVPRVISMLKKKKQ-NH2 |
| Ponericin W4 | GIWGTALKWGVKLLPKLVGMAQTKKQ-NH2 |
| Ponericin W5 | FWGALIKGAAKLIPSVVGLFKKKQ-NH2 |
| Ponericin W6 | FIGTALGIASAIPAIVKLFK-NH2 |
| Ponericin L1 | LLKELWTKMKGAGKAVLGKI-NH2 |
| Ponericin L2 | LLKELWTKIKGAGKAVLGKIKGLL-NH2 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, Q.; Patočka, J.; Kuča, K. Insect Antimicrobial Peptides, a Mini Review. Toxins 2018, 10, 461. https://0-doi-org.brum.beds.ac.uk/10.3390/toxins10110461
Wu Q, Patočka J, Kuča K. Insect Antimicrobial Peptides, a Mini Review. Toxins. 2018; 10(11):461. https://0-doi-org.brum.beds.ac.uk/10.3390/toxins10110461
Chicago/Turabian StyleWu, Qinghua, Jiří Patočka, and Kamil Kuča. 2018. "Insect Antimicrobial Peptides, a Mini Review" Toxins 10, no. 11: 461. https://0-doi-org.brum.beds.ac.uk/10.3390/toxins10110461





