Characterization of A Staphylococcal Food Poisoning Outbreak in A Workplace Canteen during the Post-Earthquake Reconstruction of Central Italy
Abstract
:1. Introduction
Background
2. Results
2.1. Epidemiological Investigation
2.2. Statistical Analysis
2.3. Laboratory Investigation
2.3.1. Sample Collection
2.3.2. Microbiological and Molecular Analysis
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Study Design
5.2. Epidemiological Investigation
5.3. Laboratory Investigation
5.3.1. Sample Collection
5.3.2. Microbiological and Molecular Analysis
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- EFSA (European Food Safety Authority) and ECDC (European Centre for Disease Prevention and control). The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2016. EFSA J. 2017, 15, e05077. [Google Scholar] [CrossRef]
- Ercoli, L.; Gallina, S.; Nia, Y.; Auvray, F.; Primavilla, A.; Guidi, F.; Pierucci, B.; Graziotti, C.; Decastelli, L.; Scuota, S. Investigation of a Staphylococcal Food Poisoning Outbreak from a Chantilly Cream Dessert, in Umbria (Italy). Foodborne Pathog. Dis. 2017, 14, 407–413. [Google Scholar] [CrossRef]
- Argudín, M.Á.; Mendoza, M.C.; Rodicio, M.R. Food Poisoning and Staphylococcus aureus Enterotoxins. Toxins 2010, 2, 1751–1773. [Google Scholar] [CrossRef] [PubMed]
- Gallina, S.; Bianchi, D.M.; Bellio, A.; Nogarol, C.; Macori, G.; Zaccaria, T.; Biorci, F.; Carraro, E.; Decastelli, L. Staphylococcal poisoning foodborne outbreak: Epidemiological investigation and strain genotyping. J. Food Prot. 2013, 76, 2093–2098. [Google Scholar] [CrossRef] [PubMed]
- Schmid, D.; Gschiel, E.; Mann, M.; Huhulescu, S.; Ruppitsch, W.; Bohm, G.; Pichler, J.; Lederer, I.; Höger, G.; Heuberger, S.; Allerberger, F. Outbreak of acute gastroenteritis in an Austrian boarding school. Euro. Surveill. 2007, 12, 224. [Google Scholar] [CrossRef] [PubMed]
- Nunes, M.M.; Caldas, E.D. Preliminary Quantitative Microbial Risk Assessment for Staphylococcus enterotoxins in fresh Minas cheese, a popular food in Brazil. Food Control 2017, 73, 524–531. [Google Scholar] [CrossRef]
- Normanno, G.; La Salandra, G.; Dambrosio, A.; Quaglia, N.C.; Corrente, M.; Parisi, A.; Santagada, G.; Firinu, A.; Crisetti, E.; Celano, G.V. Occurrence, characterization and antimicrobial resistance of enterotoxigenic Staphylococcus aureus isolated from meat and dairy products. Int. J. Food Microbiol. 2007, 115, 290–296. [Google Scholar] [CrossRef]
- Hennekinne, J.A.; Ostyn, A.; Guiller, F.; Herbin, S.; Prufer, A.L.; Dragacci, S. How Should Staphylococcal Food Poisoning Outbreaks Be Characterized. Toxins 2010, 2, 2106–2116. [Google Scholar] [CrossRef]
- Hait, J.M.; Nguyen, A.T.; Tallent, S.M. Analysis of the VIDAS® Staph Enterotoxin III (SET3) for Detection of Staphylococcal Enterotoxins G, H and I in Foods. J. AOAC Int. 2018, 101, 1482–1489. [Google Scholar] [CrossRef]
- Hennekinne, J.A.; Kerouanton, A.; Brisabois, A.; De Buyser, M.L. Discrimination of Staphylococcus aureus biotypes by pulsed-field gel electrophoresis of DNA macro-restriction fragments. J. Appl. Microbiol. 2003, 94, 321–329. [Google Scholar] [CrossRef]
- International Organization for Standardization. Microbiology of Food and Animal Feeding Stuffs—General Requirements and Guidance for Microbiological Examinations; ISO: Geneva, Switzerland, 2007. [Google Scholar]
- Guillier, L.; Bergis, H.; Guillier, F.; Noel, V.; Auvray, F.; Hennekinne, J.A. Dose-response modelling of staphylococcal enterotoxins using outbreak data. Procedia Food Sci. 2016, 7, 129–132. [Google Scholar] [CrossRef]
- Hennekinne, J.A.; Brun, V.; De Buyser, M.L.; Dupuis, A.; Ostyn, A.; Dragacci, S. Innovative Application of Mass Spectrometry for the Characterization of staphylococcal Enterotoxins Involved in Food Poisoning Outbreaks. Appl. Environ. Microb. 2009, 75, 882–884. [Google Scholar] [CrossRef] [PubMed]
- Ostyn, A.; De Buyser, M.L.; Guiller, F.; Groult, J.; Félix, B.; Salah, S.; Delmas, G.; Hennekinne, J.A. First evidence of a food poisoning outbreak due to staphylococcal enterotoxin type E, France, 2009. Eurosurveillance 2010, 15, 1–4. [Google Scholar] [CrossRef]
- Denayer, S.; Delbrassinne, L.; Nia, Y.; Botteldoorn, N. Food-Borne Outbreak Investigation and Molecular Typing: High Diversity of Staphylococcus aureus Strains and Importance of Toxin Detection. Toxins 2017, 9, 407. [Google Scholar] [CrossRef] [PubMed]
- Chiang, Y.; Liao, W.; Fan, C.; Pai, W.; Chiou, C.; Tsen, H. PCR detection of Staphylococcus aureus isolates from food-poisoning cases in Taiwan. Int. J. Food Microbiol. 2008, 121, 66–73. [Google Scholar] [CrossRef] [PubMed]
- Ciupescu, L.M.; Auvray, F.; Nicorescu, I.M.; Meheut, T.; Ciupescu, V.; Lardeux, A.L.; Tanasuica, R.; Hennekinne, J.A. Characterization of Staphylococcus aureus strains and evidence for the involvement of non-classical enterotoxin genes in food poisoning outbreaks. FEMS Microbiol. Lett. 2018, 365, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Fisher, E.L.; Otto, M.; Cheung, G.Y.C. Basis of Virulence in Enterotoxin-Mediated Staphylococcal Food Poisoning. Front Microbiol. 2018, 9, 1–18. [Google Scholar] [CrossRef]
- Mertz, D.; Frei, R.; Jaussi, B.; Tietz, A.; Stebler, C.; Flückiger, U.; Widmer, A.F. Throat swabs are necessary to reliably detect carriers of Staphylococcus aureus. Clin. Infect. Dis. 2007, 45, 475–477. [Google Scholar] [CrossRef]
- Solano, R.; Lafuente, S.; Sabate, S.; Tortajada, C.; de Olalla, P.G.; Hernando, A.V.; Caylà, J. Enterotoxin production by Staphylococcus aureus: An outbreak at a Barcelona sports club in July 2011. Food Control 2013, 33, 114–118. [Google Scholar] [CrossRef]
- World Health Organization (WHO). Foodborne Disease Outbreaks: Guidelines for Investigation and Control; WHO Library Cataloguing-in-Publication Data; WHO: Geneva, Switzerland, 2008. [Google Scholar]
- Repertorio Atti n.212. Conferenza Stato Regioni del 10 Novembre 2016: Linee guida per il controllo ufficiale ai sensi dei Regolamenti (CE) 882/2004 e 854/2004. Available online: http://www.regioni.it/ue-esteri/2016/11/29/conferenza-stato-regioni-del-10-11-2016-intesa-sul-documento-concernente-linee-guida-per-il-controllo-ufficiale-ai-sensi-dei-regolamenti-ce-8822004-e-8542004-488553/ (accessed on 11 November 2016).
- ISO 6888-2:2004. Microbiology of Food and Animal Feeding Stuffs—Horizontal Method for the Enumeration of Coagulase-Positive Staphylococci (Staphylococcus aureus and Other Species)—Technique Using Rabbit Plasma Fibrinogen Agar Medium; International Organization for Standardization: Geneva, Switzerland, 2004. [Google Scholar]
- ISO 7932:2004. Microbiology of Food and Animal Feeding Stuffs—Horizontal Method for the Enumeration of Presumptive Bacillus cereus-Colony-Count Technique at Degree 30; International Organization for Standardization: Geneva, Switzerland, 2004. [Google Scholar]
- ISO 18593:2004. Microbiology of Food and Animal Feeding Stuffs—Horizontal Methods for Sampling Techniques from Surfaces Using Contact Plates and Swabs; International Organization for Standardization: Geneva, Switzerland, 2004. [Google Scholar]
- Ostyn, A.; Guillier, F.; Prufer, A.L.; Papinaud, I.; Messio, S.; Krys, S.; Lombard, B.; Hennekinne, J.A. Intra-laboratory validation of the Ridascreen® SET Total kit for detecting staphylococcal enterotoxins SEA to SEE in cheese. Lett. Appl. Microbiol. 2011, 52, 468–474. [Google Scholar] [CrossRef]
- Ostyn, A.; Prufer, A.L.; Papinaud, I.; Hennekinne, J.A.; Assere, A.; Lombard, B. European screening method of the EURL for CPS including Staphylococcus aureus applicable to the detection of SEs in all types of food matrices including milk and milk products. 2016. Available online: http://eurl-staphylococci.anses.fr (accessed on 11 November 2016).
- Hennekinne, J.A.; Guillier, F.; Perelle, S.; De Buyser, M.L.; Dragacci, S.; Krys, S.; Lombard, B. Interlaboratory validation according to the EN ISO 16 140 Standard of the Vidas SET2 detection kit for use in official controls of staphylococcal enterotoxins in milk products. J. Appl. Microbiol. 2007, 102, 1261–1272. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Sparling, J.; Chow, B.L.; Elsayed, S.; Hussain, Z.; Church, D.L.; Gregson, D.B.; Louie, T.; Colony, J.M. New Quadriplex PCR Assay for Detection of Methicillin and Mupirocin Resistance and Simultaneous Discrimination of Staphylococcus aureus from Coagulase-Negative Staphylococci. J. Clin. Microbiol. 2004, 42, 4947–4955. [Google Scholar] [CrossRef] [PubMed]
- De Buyser, M.L.; Grout, J.; Brisabois, A.; Assere, A.; Lombard, B. Detection of Genes Encoding Staphylococcal Enterotoxins Multiplex PCR for Sea to See and Ser. Method of the CRL for Coagulase Positive Staphylococci, Including Staphylococcus aureus; Version 1. CRL CPS; AFSSA: Maisons-Alfort, France, 2009a; pp. 1–5. [Google Scholar]
- De Buyser, M.L.; Grout, J.; Brisabois, A.; Assere, A.; Lombard, B. Detection of Genes Encoding Staphylococcal Enterotoxins Multiplex PCR for Seg to Sej and Sep. Method of the CRL for Coagulase Positive Staphylococci, Including Staphylococcus aureus; Version 1. CRL CPS; AFSSA: Maisons-Alfort, France, 2009b; pp. 1–5. [Google Scholar]
- Marault, M.; Roussel, S.; Brisabois, A. Staphylococcus aureus Pulsed Field Gel Electrophoresis subtyping method. Méthode ANSES Maisons-Alfort CEB 06 (rev 04). 28 février 2011. [Google Scholar]
- Devriese, L.A. A simplified system for biotyping Staphylococcus aureus strains isolated from different animal species. J. Appl. Bacteriol. 1984, 56, 215–220. [Google Scholar] [CrossRef] [PubMed]
- Kérouanton, A.; Hennekinne, J.A.; Letertre, C.; Petit, L.; Chesneau, O.; Brisabois, A.; De Buyser, M.L. Characterization of Staphylococcus aureus strains associated with food poisoning outbreaks in France. Int. J. Food Microbiol. 2007, 115, 369–375. [Google Scholar] [CrossRef] [PubMed]
- Manzano, M.; Cocolin, L.; Cantoni, C.; Comi, G. Bacillus cereus, Bacillus Thuringensis and Bacillus mycoides differentiation using a PCR-RE technique. Int. J. Food Microbiol. 2003, 81, 249–254. [Google Scholar] [CrossRef]
- Kim, J.B.; Kim, J.M.; Park, Y.B.; Han, J.A.; Lee, S.H.; Kwak, H.S.; Hwang, I.G.; Yoon, M.H.; Lee, J.B.; Oh, D.H. Evaluation of various PCR assays for the detection of emetic toxin producing Bacillus cereus. J. Microbiol. Biotechnol. 2010, 20, 1107–1113. [Google Scholar] [PubMed]
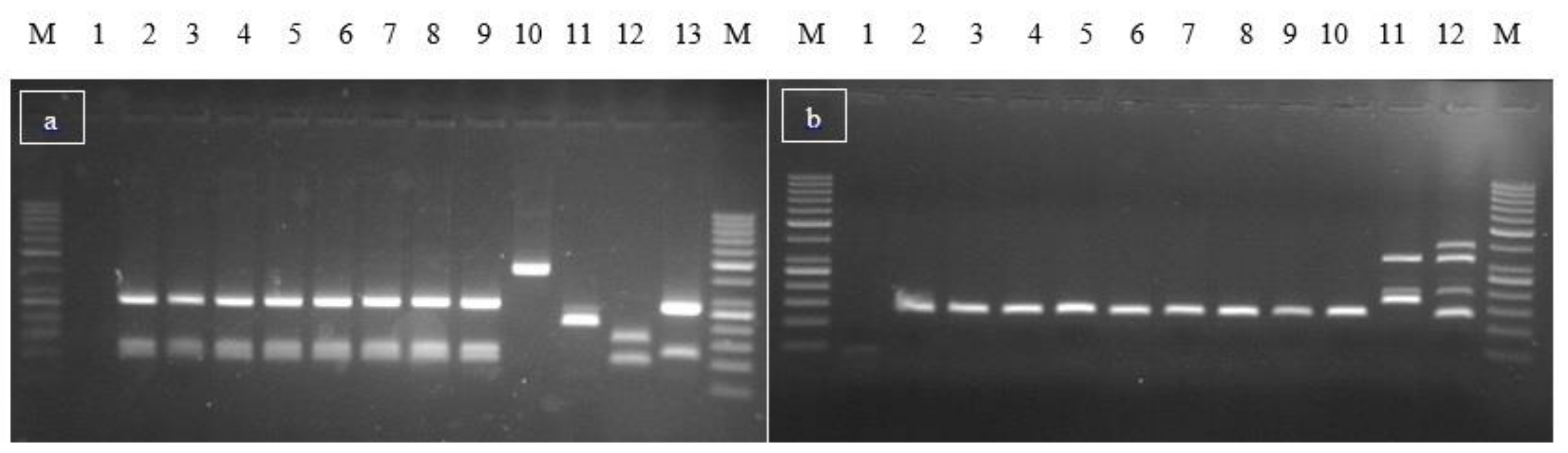
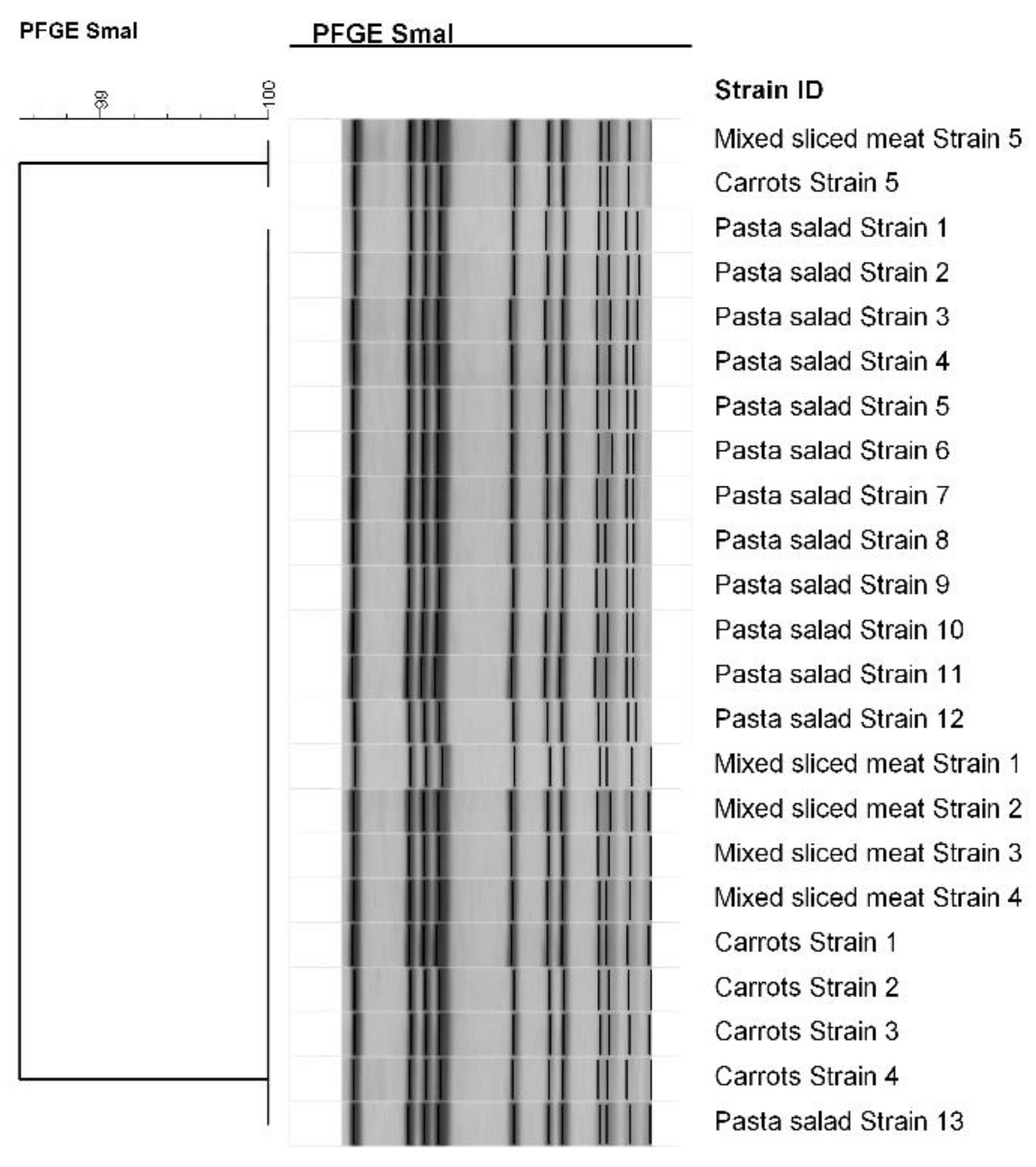
| Foods | Exposed (Ate the Specific Food) | Not Exposed (Did Not Eat the Specific Food) | RR | RA | IC 95% | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Cases | Total | AR | Cases | Total | AR | Lower | Upper | |||
| 1. Pasta salad | 26 | 36 | 72% | 0 | 0 | n.a. | n.a. | 72% | 57% | 87% |
| 2. Carrots | 11 | 16 | 68.75% | 15 | 20 | 75.00% | 0.92 | / | 0.60 | 1.31 |
| 3. Ham | 21 | 29 | 72.41% | 5 | 7 | 71.43% | 1.01 | / | 0.60 | 1.71 |
| 4. Loin | 18 | 25 | 72.00% | 8 | 11 | 72.73% | 0.99 | / | 0.64 | 1.53 |
| 5. Salami | 22 | 31 | 70.97% | 4 | 5 | 80.00% | 0.89 | / | 0.54 | 1.45 |
| Food sample | Sample Unit Number | CPS Enumeration (CFU/g) | Staphylococcal Enterotoxin (Detection) | Staphylococcal Enterotoxins (Quantification) | Bacillus cereus Enumeration (CFU/g) |
|---|---|---|---|---|---|
| Mixed sliced meat | Single unit | 1500 | Not done | / | <100 |
| Pasta salad | Single unit | 99,000,000 | Detected | SEA 0.033 ng/g; SEB < LoQ; SEC < LoD; SED 0.052 ng/g | >150,000 |
| Boiled carrots | Single unit | 610 | Not found | / | <100 |
| Baked ham | 1 | <10 | Not found | / | 100 |
| 2 | <10 | Not found | / | <100 | |
| 3 | <10 | Not found | / | Detected <400 | |
| 4 | <10 | Not found | / | <100 | |
| 5 | <10 | Not found | / | <100 | |
| Emmental cheese | 1 | <10 | Not found | / | <100 |
| 2 | <10 | Not found | / | <100 | |
| 3 | <10 | Not found | / | <100 | |
| 4 | <10 | Not found | / | <100 | |
| 5 | <10 | Not found | / | <100 | |
| Provolone cheese | 1 | <10 | Not found | / | <100 |
| 2 | <10 | Not found | / | Detected <400 | |
| 3 | <10 | Not found | / | <100 | |
| 4 | <10 | Not found | / | <100 | |
| 5 | <10 | Not found | / | <100 | |
| Mixed cheese | 1 | <10 | Not found | / | <100 |
| 2 | <10 | Not found | / | <100 | |
| 3 | <10 | Not found | / | <100 | |
| 4 | <10 | Not found | / | <100 | |
| 5 | <10 | Not found | / | <100 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guidi, F.; Duranti, A.; Gallina, S.; Nia, Y.; Petruzzelli, A.; Romano, A.; Travaglini, V.; Olivastri, A.; Calvaresi, V.; Decastelli, L.; et al. Characterization of A Staphylococcal Food Poisoning Outbreak in A Workplace Canteen during the Post-Earthquake Reconstruction of Central Italy. Toxins 2018, 10, 523. https://0-doi-org.brum.beds.ac.uk/10.3390/toxins10120523
Guidi F, Duranti A, Gallina S, Nia Y, Petruzzelli A, Romano A, Travaglini V, Olivastri A, Calvaresi V, Decastelli L, et al. Characterization of A Staphylococcal Food Poisoning Outbreak in A Workplace Canteen during the Post-Earthquake Reconstruction of Central Italy. Toxins. 2018; 10(12):523. https://0-doi-org.brum.beds.ac.uk/10.3390/toxins10120523
Chicago/Turabian StyleGuidi, Fabrizia, Anna Duranti, Silvia Gallina, Yacine Nia, Annalisa Petruzzelli, Angelo Romano, Valeria Travaglini, Alberto Olivastri, Vincenzo Calvaresi, Lucia Decastelli, and et al. 2018. "Characterization of A Staphylococcal Food Poisoning Outbreak in A Workplace Canteen during the Post-Earthquake Reconstruction of Central Italy" Toxins 10, no. 12: 523. https://0-doi-org.brum.beds.ac.uk/10.3390/toxins10120523





