Determination of the Role of Microcystis aeruginosa in Toxin Generation Based on Phosphoproteomic Profiles
Abstract
:1. Introduction
2. Results
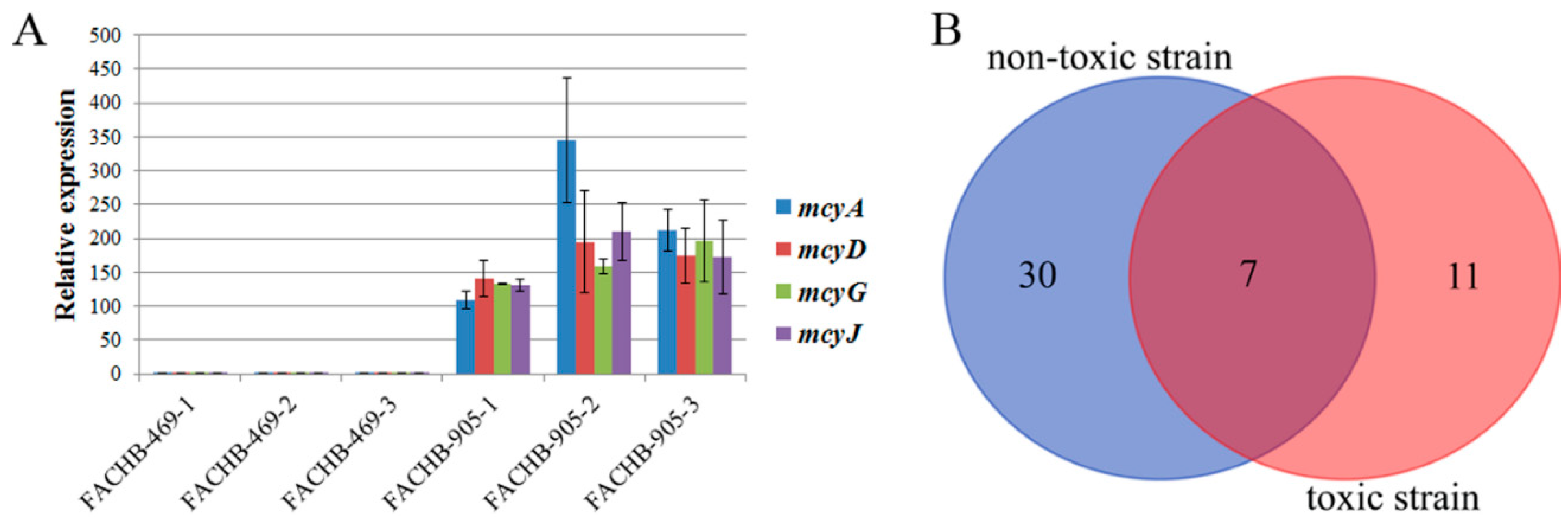
2.1. Global Phosphoproteome Characterization of Toxic and Non-Toxic M. aeruginosa Strains
2.2. Differences in Protein Phosphorylation Abundance and Function between the Toxic and Non-Toxic M. aeruginosa Strains
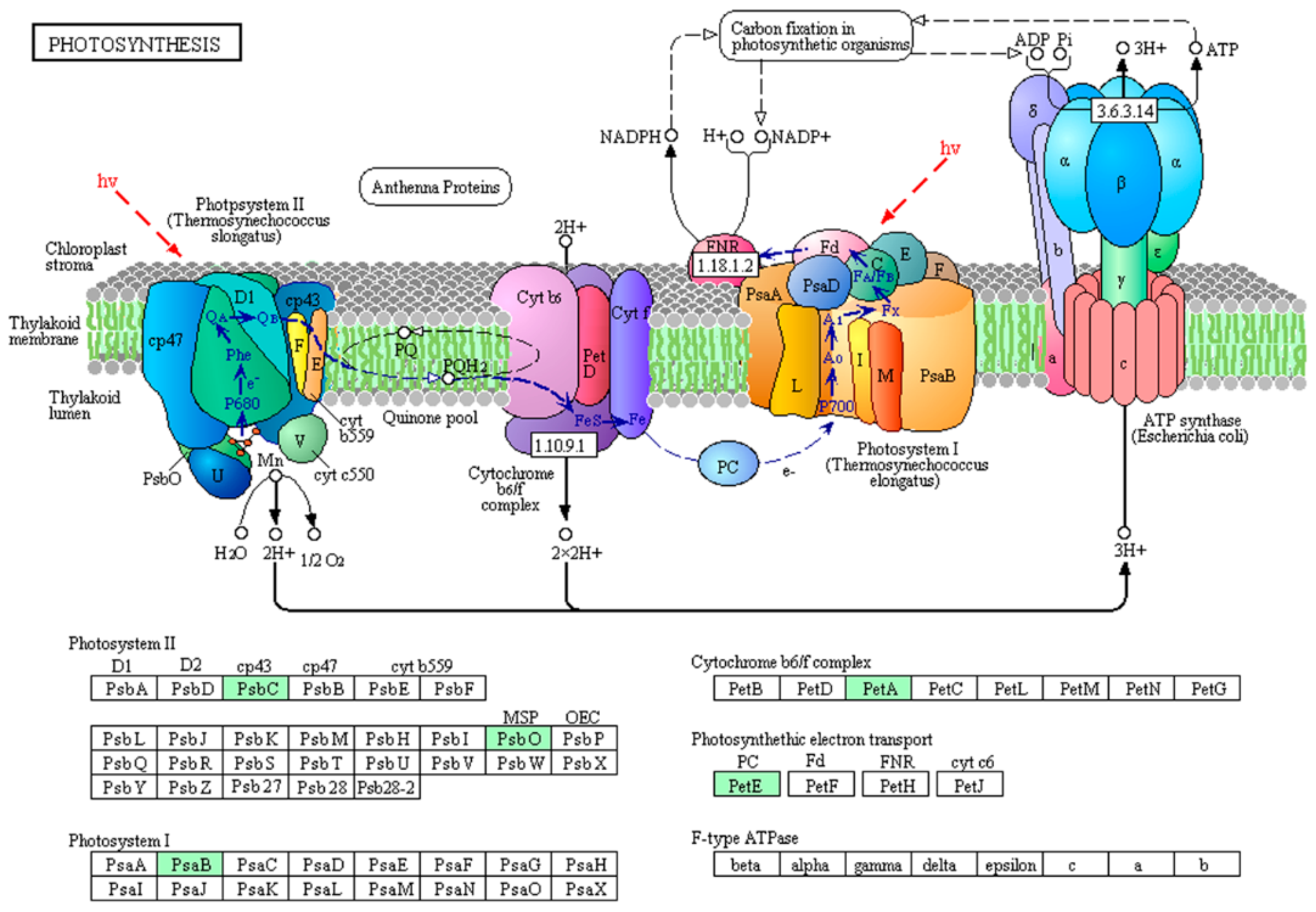
2.3. Protein Interaction Network of the Phosphorylated Proteins in the Toxic and Non-Toxic Strains
3. Discussion
4. Materials and Methods
4.1. Microcystis Aeruginosa Sample Preparation
4.2. Real-Time PCR (qPCR) Analysis
4.3. Protein Extraction and Digestion
4.4. Affinity Enrichment of the Phosphorylated Peptides
4.5. LC-MS/MS Analysis
4.6. Database Search and Quantification
4.7. Bioinformatics Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Dagan, T.; Roettger, M.; Stucken, K.; Landan, G.; Koch, R.; Major, P.; Gould, S.B.; Goremykin, V.V.; Rippka, R.; Tandeau de Marsac, N.; et al. Genomes of Stigonematalean cyanobacteria (subsection V) and the evolution of oxygenic photosynthesis from prokaryotes to plastids. Genome Biol. Evol. 2013, 5, 31–44. [Google Scholar] [CrossRef] [PubMed]
- Baldia, S.F.; Conaco, M.C.G.; Nishijima, T.; Imanishi, S.; Harada, K.I. Microcystin production during algal bloom occurrence in Laguna de Bay, the Philippines. Fish. Sci. 2003, 69, 110–116. [Google Scholar] [CrossRef]
- Sinha, R.; Pearson, L.A.; Davis, T.W.; Burford, M.A.; Orr, P.T.; Neilan, B.A. Increased incidence of Cylindrospermopsis raciborskii in temperate zones—Is climate change responsible? Water Res. 2012, 46, 1408–1419. [Google Scholar] [CrossRef] [PubMed]
- Fiore, M.F.; Genuario, D.B.; da Silva, C.S.; Shishido, T.K.; Moraes, L.A.; Cantusio Neto, R.; Silva-Stenico, M.E. Microcystin production by a freshwater spring cyanobacterium of the genus Fischerella. Toxicon 2009, 53, 754–761. [Google Scholar] [CrossRef] [PubMed]
- Rantala, A.; Fewer, D.P.; Hisbergues, M.; Rouhiainen, L.; Vaitomaa, J.; Borner, T.; Sivonen, K. Phylogenetic evidence for the early evolution of microcystin synthesis. Proc. Natl. Acad. Sci. USA 2004, 101, 568–573. [Google Scholar] [CrossRef] [PubMed]
- Kaebernick, M.; Neilan, B.A. Ecological and molecular investigations of cyanotoxin production. FEMS Microbiol. Ecol. 2001, 35, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Tonk, L.; Visser, P.M.; Christiansen, G.; Dittmann, E.; Snelder, E.O.; Wiedner, C.; Mur, L.R.; Huisman, J. The microcystin composition of the cyanobacterium Planktothrix agardhii changes toward a more toxic variant with increasing light intensity. Appl. Environ. Microbiol. 2005, 71, 5177–5181. [Google Scholar] [CrossRef] [PubMed]
- Bagu, J.R.; Sykes, B.D.; Craig, M.M.; Holmes, C.F. A molecular basis for different interactions of marine toxins with protein phosphatase-1. Molecular models for bound motuporin, microcystins, okadaic acid, and calyculin A. J. Biol. Chem. 1997, 272, 5087–5097. [Google Scholar] [CrossRef] [PubMed]
- Welker, M.; von Dohren, H. Cyanobacterial peptides—Nature’s own combinatorial biosynthesis. FEMS Microbiol. Rev. 2006, 30, 530–563. [Google Scholar] [CrossRef] [PubMed]
- Carmichael, W.W. Health effects of toxin-producing cyanobacteria: “The CyanoHABs”. Hum. Ecol. Risk Assess. 2001, 7, 1393–1407. [Google Scholar] [CrossRef]
- Soares, R.M.; Yuan, M.; Servaites, J.C.; Delgado, A.; Maglhaes, V.F.; Hilborn, E.D.; Carmichael, W.W.; Azevedol, S.M.F.O. Sublethal exposure from microcystins to renal insufficiency patients in Rio de Janeiro, Brazil. Environ. Toxicol. 2006, 21, 95–103. [Google Scholar] [CrossRef] [PubMed]
- Azevedo, S.M.; Carmichael, W.W.; Jochimsen, E.M.; Rinehart, K.L.; Lau, S.; Shaw, G.R.; Eaglesham, G.K. Human intoxication by microcystins during renal dialysis treatment in Caruaru-Brazil. Toxicology 2002, 181–182, 441–446. [Google Scholar] [CrossRef]
- Kameyama, K.; Sugiura, N.; Inamori, Y.; Maekawa, T. Characteristics of microcystin production in the cell cycle of Microcystis viridis. Environ. Toxicol. 2004, 19, 20–25. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.J.; Jang, M.H.; Kim, H.S.; Yoon, B.D.; Oh, H.M. Variation of microcystin content of microcystis aeruginosa relative to medium N:P ratio and growth stage. J. Appl. Microbiol. 2000, 89, 323–329. [Google Scholar] [CrossRef] [PubMed]
- Dittmann, E.; Neilan, B.A.; Erhard, M.; von Döhren, H.; Börner, T. Insertional mutagenesis of a peptide synthetase gene that is responsible for hepatotoxin production in the cyanobacteriumMicrocystis aeruginosaPCC 7806. Mol. Microbiol. 1997, 26, 779–787. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tillett, D.; Dittmann, E.; Erhard, M.; von Dohren, H.; Borner, T.; Neilan, B.A. Structural organization of microcystin biosynthesis in Microcystis aeruginosa PCC7806: An integrated peptide-polyketide synthetase system. Chem. Biol. 2000, 7, 753–764. [Google Scholar] [CrossRef]
- Schatz, D.; Keren, Y.; Hadas, O.; Carmeli, S.; Sukenik, A.; Kaplan, A. Ecological implications of the emergence of non-toxic subcultures from toxic Microcystis strains. Environ. Microbiol. 2005, 7, 798–805. [Google Scholar] [CrossRef] [PubMed]
- Kehr, J.C.; Zilliges, Y.; Springer, A.; Disney, M.D.; Ratner, D.D.; Bouchier, C.; Seeberger, P.H.; de Marsac, N.T.; Dittmann, E. A mannan binding lectin is involved in cell-cell attachment in a toxic strain of Microcystis aeruginosa. Mol. Microbiol. 2006, 59, 893–906. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dittmann, E.; Erhard, M.; Kaebernick, M.; Scheler, C.; Neilan, B.A.; von Dohren, H.; Borner, T. Altered expression of two light-dependent genes in a microcystin-lacking mutant of Microcystis aeruginosa PCC 7806. Microbiology 2001, 147, 3113–3119. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaneko, T.; Nakajima, N.; Okamoto, S.; Suzuki, I.; Tanabe, Y.; Tamaoki, M.; Nakamura, Y.; Kasai, F.; Watanabe, A.; Kawashima, K.; et al. Complete genomic structure of the bloom-forming toxic cyanobacterium Microcystis aeruginosa NIES-843. DNA Res. 2007, 14, 247–256. [Google Scholar] [CrossRef] [PubMed]
- Alexova, R.; Haynes, P.A.; Ferrari, B.C.; Neilan, B.A. Comparative protein expression in different strains of the bloom-forming cyanobacterium Microcystis aeruginosa. Mol. Cell. Proteom. 2011, 10, M110.003749. [Google Scholar] [CrossRef] [PubMed]
- Tonietto, A.; Petriz, B.A.; Araujo, W.C.; Mehta, A.; Magalhaes, B.S.; Franco, O.L. Comparative proteomics between natural Microcystis isolates with a focus on microcystin synthesis. Proteome Sci. 2012, 10, 38. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alexova, R.; Dang, T.C.; Fujii, M.; Raftery, M.J.; Waite, T.D.; Ferrari, B.C.; Neilan, B.A. Specific global responses to N and Fe nutrition in toxic and non-toxic Microcystis aeruginosa. Environ. Microbiol. 2016, 18, 401–413. [Google Scholar] [CrossRef] [PubMed]
- Yue, D.M.; Peng, Y.K.; Yin, Q.; Xiao, L. Proteomic analysis of Microcystis aeruginosa in response to nitrogen and phosphorus starvation. J. Appl. Phycol. 2015, 27, 1195–1204. [Google Scholar] [CrossRef]
- Rogers, L.D.; Foster, L.J. Phosphoproteomics—Finally fulfilling the promise? Mol. Biosyst. 2009, 5, 1122–1129. [Google Scholar] [CrossRef] [PubMed]
- Soufi, B.; Jers, C.; Hansen, M.E.; Petranovic, D.; Mijakovic, I. Insights from site-specific phosphoproteomics in bacteria. Biochim. Biophys. Acta 2008, 1784, 186–192. [Google Scholar] [CrossRef] [PubMed]
- Sobczyk, A.; Schyns, G.; Tandeau de Marsac, N.; Houmard, J. Transduction of the light signal during complementary chromatic adaptation in the cyanobacterium Calothrix sp. PCC 7601: DNA-binding proteins and modulation by phosphorylation. EMBO J. 1993, 12, 997–1004. [Google Scholar] [PubMed]
- Baniulis, D.; Yamashita, E.; Whitelegge, J.P.; Zatsman, A.I.; Hendrich, M.P.; Hasan, S.S.; Ryan, C.M.; Cramer, W.A. Structure-function, stability, and chemical modification of the cyanobacterial cytochrome b6f complex from Nostoc sp. PCC 7120. J. Biol. Chem. 2009, 284, 9861–9869. [Google Scholar] [CrossRef] [PubMed]
- Los, D.A.; Zorina, A.; Sinetova, M.; Kryazhov, S.; Mironov, K.; Zinchenko, V.V. Stress sensors and signal transducers in cyanobacteria. Sensors 2010, 10, 2386–2415. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.C.; Jang, J.; Sakr, S.; Wang, L. Protein phosphorylation on Ser, Thr and Tyr residues in cyanobacteria. J. Mol. Microbiol. Biotechnol. 2005, 9, 154–166. [Google Scholar] [CrossRef] [PubMed]
- Forchhammer, K. Global carbon/nitrogen control by PII signal transduction in cyanobacteria: From signals to targets. FEMS Microbiol. Rev. 2004, 28, 319–333. [Google Scholar] [CrossRef] [PubMed]
- Forchhammer, K. P-II signal transducers: Novel functional and structural insights. Trends Microbiol. 2008, 16, 65–72. [Google Scholar] [CrossRef] [PubMed]
- Piven, I.; Ajlani, G.; Sokolenko, A. Phycobilisome linker proteins are phosphorylated in Synechocystis sp. PCC 6803. J. Biol. Chem. 2005, 280, 21667–21672. [Google Scholar] [CrossRef] [PubMed]
- Harrison, M.A.; Tsinoremas, N.F.; Allen, J.F. Cyanobacterial thylakoid membrane-proteins are reversibly phosphorylated under plastoquinone-reducing conditions invitro. FEBS Lett. 1991, 282, 295–299. [Google Scholar] [CrossRef]
- Nishizawa, T.; Asayama, M.; Fujii, K.; Harada, K.; Shirai, M. Genetic analysis of the peptide synthetase genes for a cyclic heptapeptide microcystin in Microcystis spp. J. Biochem. 1999, 126, 520–529. [Google Scholar] [CrossRef] [PubMed]
- Nishizawa, T.; Ueda, A.; Asayama, M.; Fujii, K.; Harada, K.; Ochi, K.; Shirai, M. Polyketide synthase gene coupled to the peptide synthetase module involved in the biosynthesis of the cyclic heptapeptide microcystin. J. Biochem. 2000, 127, 779–789. [Google Scholar] [CrossRef] [PubMed]
- Willems, P.H.; Rossignol, R.; Dieteren, C.E.; Murphy, M.P.; Koopman, W.J. Redox homeostasis and mitochondrial dynamics. Cell Metab. 2015, 22, 207–218. [Google Scholar] [CrossRef] [PubMed]
- Jansson, S. The light-harvesting chlorophyll a/b-binding proteins. Biochim. Biophys. Acta 1994, 1184, 1–19. [Google Scholar] [CrossRef]
- Popelkova, H.; Yocum, C.F. PsbO, the manganese-stabilizing protein: Analysis of the structure-function relations that provide insights into its role in photosystem II. J. Photochem. Photobiol. B 2011, 104, 179–190. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, M.; Semchonok, D.A.; Webber-Birungi, M.T.; Ehira, S.; Kondo, K.; Narikawa, R.; Ohmori, M.; Boekema, E.J.; Ikeuchi, M. Attachment of phycobilisomes in an antenna-photosystem I supercomplex of cyanobacteria. Proc. Natl. Acad. Sci. USA 2014, 111, 2512–2517. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.Q.; Jiang, H.B.; Zhang, R.; Qiu, B.S. Inactivation of the petE gene encoding plastocyanin causes different photosynthetic responses in cyanobacterium Synechocystis PCC 6803 under light-dark photoperiod and continuous light conditions. FEMS Microbiol. Lett. 2013, 341, 106–114. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goss, T.; Hanke, G. The end of the line: Can ferredoxin and ferredoxin NADP(H) oxidoreductase determine the fate of photosynthetic electrons? Curr. Protein Pept. Sci. 2014, 15, 385–393. [Google Scholar] [CrossRef] [PubMed]
- Ubersax, J.A.; Ferrell, J.E., Jr. Mechanisms of specificity in protein phosphorylation. Nat. Rev. Mol. Cell Biol. 2007, 8, 530–541. [Google Scholar] [CrossRef] [PubMed]
- Van Riper, S.K.; de Jong, E.P.; Carlis, J.V.; Griffin, T.J. Mass spectrometry-based proteomics: Basic principles and emerging technologies and directions. Adv. Exp. Med. Biol. 2013, 990, 1–35. [Google Scholar] [PubMed]
- Mikkat, S.; Fulda, S.; Hagemann, M. A 2D gel electrophoresis-based snapshot of the phosphoproteome in the cyanobacterium Synechocystis sp. strain PCC 6803. Microbiology 2014, 160, 296–306. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.G.; Kwon, J.; Eom, C.Y.; Kang, Y.M.; Roh, S.W.; Lee, K.B.; Choi, J.S. Directed analysis of cyanobacterial membrane phosphoproteome using stained phosphoproteins and titanium-enriched phosphopeptides. J. Microbiol. 2015, 53, 279–287. [Google Scholar] [CrossRef] [PubMed]
- Rippka, R.; Stanier, R.Y.; Deruelles, J.; Herdman, M.; Waterbury, J.B. Generic assignments, strain histories and properties of pure cultures of cyanobacteria. Microbiology 1979, 111, 1–61. [Google Scholar] [CrossRef]
- Wisniewski, J.R. Quantitative evaluation of Filter Aided Sample Preparation (FASP) and multienzyme digestion FASP protocols. Anal. Chem. 2016, 88, 5438–5443. [Google Scholar] [CrossRef] [PubMed]
- Paulo, J.A.; O’Connell, J.D.; Everley, R.A.; O’Brien, J.; Gygi, M.A.; Gygi, S.P. Quantitative mass spectrometry-based multiplexing compares the abundance of 5000 S. cerevisiae proteins across 10 carbon sources. J. Proteom. 2016, 148, 85–93. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tyanova, S.; Temu, T.; Carlson, A.; Sinitcyn, P.; Mann, M.; Cox, J. Visualization of LC-MS/MS proteomics data in MaxQuant. Proteomics 2015, 15, 1453–1456. [Google Scholar] [CrossRef] [PubMed] [Green Version]




| Protein ID | Phosphorylated Peptide | GO ID and GO Name |
|---|---|---|
| L8P227 | FGDVS(1)GIVR AWS(0.026)AGT(0.487)S(0.487)VLD | Unknown function |
| B0JVG8 | LNTS(0.003)S(0.997)PFNIK | GO:0043167 ion binding |
| S3JUK9 | S(0.448)ALS(0.544)DS(0.008)NIDANQSSSQR | Unknown function |
| I4IM84 | T(0.004)S(0.004)S(0.049)Y(0.944)FGLETEENK | Unknown function |
| S3K9U2 | IT(0.035)S(0.959)NAS(0.006)TIVANAAR IT(0.022)S(0.048)NAS(0.266)T(0.664)IVANAAR | GO:0015979 photosynthesis GO:0006464 cellular protein modification process GO:0009579 thylakoid GO:0043234 protein complex |
| A8YG01 | GS(1)EYTVEFLQK | GO:0030234 enzyme regulator activity GO:0009058 biosynthetic process GO:0034641 cellular nitrogen compound metabolic process |
| L8NSK3 | AAAT(1)ELGVPAADIPTSTSR | Unknown function |
| GO ID and GO Name | Protein ID | Phosphorylated Peptide | Protein Description |
|---|---|---|---|
| GO:0042592 homeostatic process | L8NNX1 | S(1)EVPAVTDANFK | Thioredoxin: cell redox homeostasis |
| GO:0016853 isomerase activity | L8P1T1 | SENALGAIVLT(0.033)AS(0.967)HNPAK | Phosphoglucomutase/phosphomannomutase |
| GO:0016301 kinase activity | A8YB46 | S(0.328)S(0.328)T(0.328)PQKPT(0.013)VS(0.003)PAVVIQPNSGEK | Serine/threonine-protein kinase C |
| GO:0003924 GTPase activity GO:0008135 translation factor activity, RNA binding | S3IW37 | T(1)IGSGVISK | Elongation factor Tu |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qu, J.; Shen, L.; Zhao, M.; Li, W.; Jia, C.; Zhu, H.; Zhang, Q. Determination of the Role of Microcystis aeruginosa in Toxin Generation Based on Phosphoproteomic Profiles. Toxins 2018, 10, 304. https://0-doi-org.brum.beds.ac.uk/10.3390/toxins10070304
Qu J, Shen L, Zhao M, Li W, Jia C, Zhu H, Zhang Q. Determination of the Role of Microcystis aeruginosa in Toxin Generation Based on Phosphoproteomic Profiles. Toxins. 2018; 10(7):304. https://0-doi-org.brum.beds.ac.uk/10.3390/toxins10070304
Chicago/Turabian StyleQu, Jiangqi, Liping Shen, Meng Zhao, Wentong Li, Chengxia Jia, Hua Zhu, and Qingjing Zhang. 2018. "Determination of the Role of Microcystis aeruginosa in Toxin Generation Based on Phosphoproteomic Profiles" Toxins 10, no. 7: 304. https://0-doi-org.brum.beds.ac.uk/10.3390/toxins10070304





