Application of Metabolomic Tools for Studying Low Molecular-Weight Fraction of Animal Venoms and Poisons
Abstract
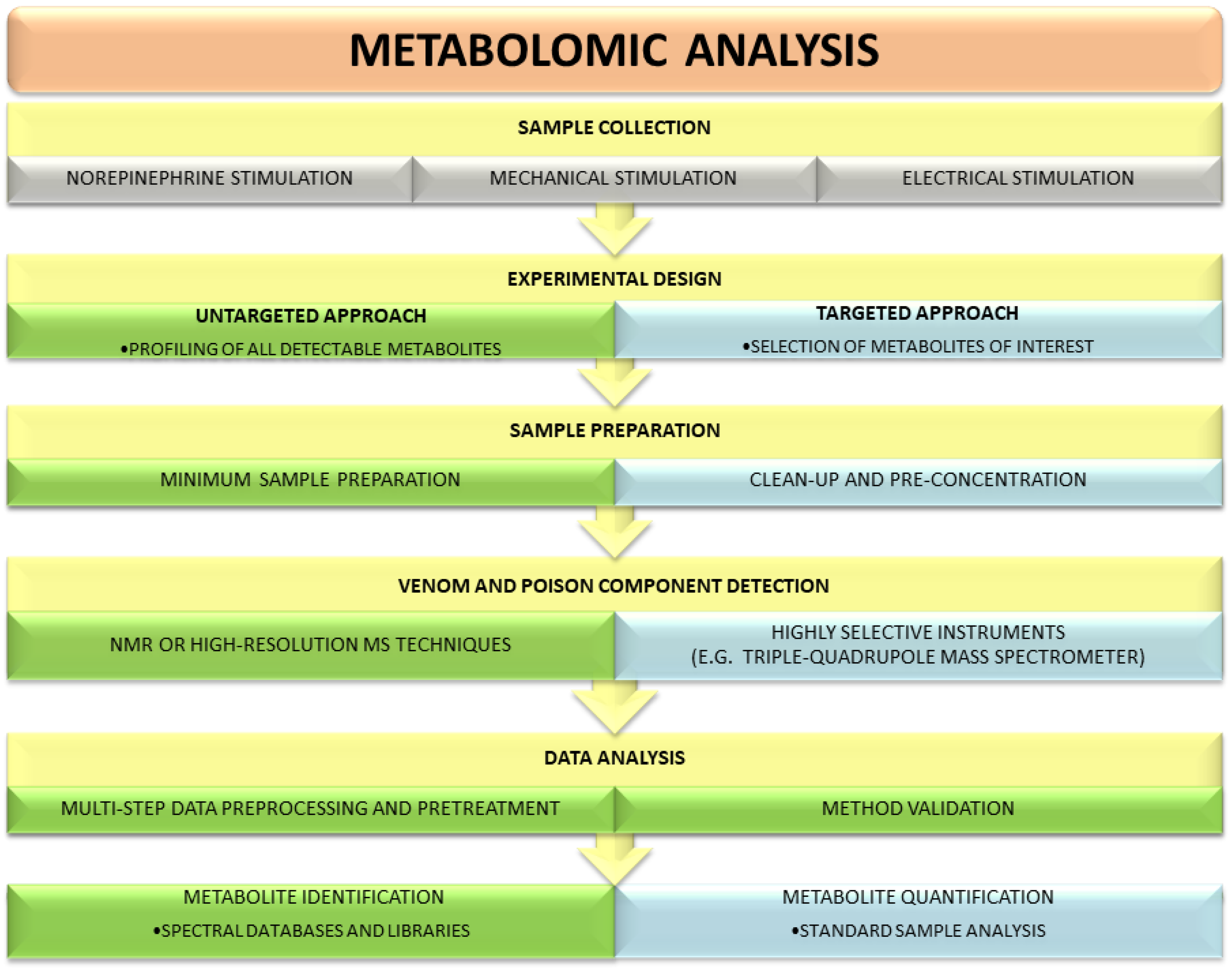
:1. Introduction
2. Aims and Methods
3. Metabolomics Strategies Used for the Analysis of Poisons and Venoms
3.1. Untargeted Metabolomic Studies
3.2. Targeted Metabolomic Studies
4. Biological Significance and Potential Applications of Small Molecule Components of Poisons and Venoms
5. Challenges and Future Perspectives
6. Concluding Remarks
Author Contributions
Funding
Conflicts of Interest
References
- von Reumont, B.M.; Campbell, L.I.; Jenner, R.A. Quo Vadis venomics? A roadmap to neglected venomous invertebrates. Toxins 2014, 6, 3488–3551. [Google Scholar] [CrossRef] [PubMed]
- Sunagar, K.; Morgenstern, D.; Reitzel, A.M.; Moran, Y. Ecological venomics: How genomics, transcriptomics and proteomics can shed new light on the ecology and evolution of venom. J. Proteom. 2016, 135, 62–72. [Google Scholar] [CrossRef] [PubMed]
- Casewell, N.R.; Wüster, W.; Vonk, F.J.; Harrison, R.A.; Fry, B.G. Complex cocktails: The evolutionary novelty of venoms. Trends Ecol. Evol. 2013, 28, 219–229. [Google Scholar] [CrossRef] [PubMed]
- Utkin, Y.N. Modern trends in animal venom research-omics and nanomaterials. World J. Biol. Chem. 2017, 8, 4. [Google Scholar] [CrossRef] [PubMed]
- Tan, N.H.; Wong, K.Y.; Tan, C.H. Venomics of Naja sputatrix, the Javan spitting cobra: A short neurotoxin-driven venom needing improved antivenom neutralization. J. Proteom. 2017, 157, 18–32. [Google Scholar] [CrossRef] [PubMed]
- Juárez, P.; Sanz, L.; Calvete, J.J. Snake venomics: Characterization of protein families in Sistrurus barbouri venom by cysteine mapping, N-terminal sequencing, and tandem mass spectrometry analysis. Proteomics 2004, 4, 327–338. [Google Scholar] [CrossRef] [PubMed]
- Bazaa, A.; Marrakchi, N.; El Ayeb, M.; Sanz, L.; Calvete, J.J. Snake venomics: Comparative analysis of the venom proteomes of the Tunisian snakes Cerastes cerastes, Cerastes vipera and Macrovipera lebetina. Proteomics 2005, 5, 4223–4235. [Google Scholar] [CrossRef] [PubMed]
- Ménez, A.; Stöcklin, R.; Mebs, D. ‘Venomics’ or: The venomous systems genome project. Toxicon 2006, 47, 255–259. [Google Scholar] [CrossRef] [PubMed]
- Biass, D.; Violette, A.; Hulo, N.; Lisacek, F.; Favreau, P.; Stöcklin, R. Uncovering intense protein diversification in a cone snail venom gland using an integrative venomics approach. J. Proteome Res. 2015, 14, 628–638. [Google Scholar] [CrossRef] [PubMed]
- Viala, V.L.; Hildebrand, D.; Trusch, M.; Fucase, T.M.; Sciani, J.M.; Pimenta, D.C.; Arni, R.K.; Schlüter, H.; Betzel, C.; Mirtschin, P.; et al. Venomics of the Australian eastern brown snake (Pseudonaja textilis): Detection of new venom proteins and splicing variants. Toxicon 2015, 107, 252–265. [Google Scholar] [CrossRef] [PubMed]
- Nicholson, J.K.; Lindon, J.C.; Holmes, E. “Metabonomics”: Understanding the metabolic responses of living systems to pathophysiological stimuli via multivariate statistical analysis of biological NMR spectroscopic data. Xenobiotica 1999, 29, 1181–1189. [Google Scholar] [CrossRef] [PubMed]
- Liesenfeld, D.B.; Habermann, N.; Owen, R.W.; Scalbert, A.; Ulrich, C.M. Review of mass spectrometry-based metabolomics in cancer research. Cancer Epidemiol. Biomark. Prev. 2013, 22, 2182–2201. [Google Scholar] [CrossRef] [PubMed]
- Sandlers, Y. The future perspective: Metabolomics in laboratory medicine for inborn errors of metabolism. Transl. Res. 2017, 189, 65–75. [Google Scholar] [CrossRef] [PubMed]
- Gil de la Fuente, A.; Grace Armitage, E.; Otero, A.; Barbas, C.; Godzien, J. Differentiating signals to make biological sense—A guide through databases for MS-based non-targeted metabolomics. Electrophoresis 2017, 38, 2242–2256. [Google Scholar] [CrossRef] [PubMed]
- Matysiak, J.; Dereziński, P.; Klupczyńska, A.; Matysiak, J.; Kaczmarek, E.; Kokot, Z.J. Effects of a honeybee sting on the serum free amino acid profile in humans. PLoS ONE 2014, 9, e103533. [Google Scholar] [CrossRef] [PubMed]
- Arjmand, M.; Akbari, Z.; Taghizadeh, N.; Shahbazzadeh, D.; Zamani, Z. NMR-based metabonomics survey in rats envenomed by Hemiscorpius lepturus venom. Toxicon 2015, 94, 16–22. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Zhang, J.; Chen, Y.; Li, Z.; Nie, H.; Peng, W.; Su, S. Altered Serum Metabolite Profiling and Relevant Pathway Analysis in Rats Stimulated by Honeybee Venom: New Insight into Allergy to Honeybee Venom. J. Agric. Food Chem. 2018, 66, 871–880. [Google Scholar] [CrossRef] [PubMed]
- Siebert, A.L.; Wright, J.; Martinson, E.; Wheeler, D.; Werren, J.H. Parasitoid venom induces metabolic cascades in fly hosts. Metabolomics 2015, 11, 350–366. [Google Scholar] [CrossRef]
- Ma, H.; Niu, H.; Cao, Q.; Zhou, J.; Gong, Y.; Zhu, Z.; Lv, X.; Di, L.; Qian, D.; Wu, Q.; et al. Metabolomics method based on ultra high performance liquid chromatography with time-of-flight mass spectrometry to analyze toxins in fresh and dried toad venom. J. Sep. Sci. 2016, 39, 4681–4687. [Google Scholar] [CrossRef] [PubMed]
- Meng, Q.; Yau, L.F.; Lu, J.G.; Wu, Z.Z.; Zhang, B.X.; Wang, J.R.; Jiang, Z.H. Chemical profiling and cytotoxicity assay of bufadienolides in toad venom and toad skin. J. Ethnopharmacol. 2016, 187, 74–82. [Google Scholar] [CrossRef] [PubMed]
- Gao, H.; Zehl, M.; Leitner, A.; Wu, X.; Wang, Z.; Kopp, B. Comparison of toad venoms from different Bufo species by HPLC and LC-DAD-MS/MS. J. Ethnopharmacol. 2010, 131, 368–376. [Google Scholar] [CrossRef] [PubMed]
- Sciani, J.M.; Angeli, C.B.; Antoniazzi, M.M.; Jared, C.; Pimenta, D.C. Differences and similarities among parotoid macrogland secretions in south American toads: A preliminary biochemical delineation. Sci. World J. 2013, 2013. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Wang, L.; Ye, W.C.; Yao, Z.P. In vivo and real-time monitoring of secondary metabolites of living organisms by mass spectrometry. Sci. Rep. 2013, 3, 2104. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Gong, Y.; Ma, H.; Wang, H.; Qian, D.; Wen, H.; Liu, R.; Duan, J.; Wu, Q. Effect of drying methods on the free and conjugated bufadienolide content in toad venom determined by ultra-performance liquid chromatography-triple quadrupole mass spectrometry coupled with a pattern recognition approach. J. Pharm. Biomed. Anal. 2015, 114, 482–487. [Google Scholar] [CrossRef] [PubMed]
- Cavalcante, I.D.; Antoniazzi, M.M.; Jared, C.; Pires, O.R.; Sciani, J.M.; Pimenta, D.C. Venomics analyses of the skin secretion of Dermatonotus muelleri: Preliminary proteomic and metabolomic profiling. Toxicon 2017, 130, 127–135. [Google Scholar] [CrossRef] [PubMed]
- Mariano, D.O.C.; Yamaguchi, L.F.; Jared, C.; Antoniazzi, M.M.; Sciani, J.M.; Kato, M.J.; Pimenta, D.C. Pipa carvalhoi skin secretion profiling: Absence of peptides and identification of kynurenic acid as the major constitutive component. Comp. Biochem. Physiol. Part C Toxicol. Pharmacol. 2015, 167, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Schroeder, F.C.; Taggi, A.E.; Gronquist, M.; Malik, R.U.; Grant, J.B.; Eisner, T.; Meinwald, J. NMR-spectroscopic screening of spider venom reveals sulfated nucleosides as major components for the brown recluse and related species. Proc. Natl. Acad. Sci. USA 2008, 105, 14283–14287. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tzouros, M.; Manov, N.; Bienz, S.; Bigler, L. Tandem mass spectrometric investigation of acylpolyamines of spider venoms and their 15N-labeled derivatives. J. Am. Soc. Mass Spectrom. 2004, 15, 1636–1643. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tzouros, M.; Chesnov, S.; Bigler, L.; Bienz, S. A template approach for the characterization of linear polyamines and derivatives in spider venom. Eur. J. Mass Spectrom. 2013, 19, 57–69. [Google Scholar] [CrossRef]
- Hisada, M.; Fujita, T.; Naoki, H.; Itagaki, Y.; Irie, H.; Miyashita, M.; Nakajima, T. Structures of spider toxins: Hydroxyindole-3-acetylpolyamines and a new generalized structure of type-E compounds obtained from the venom of the Joro spider, Nephila clavata. Toxicon 1998, 36, 1115–1125. [Google Scholar] [CrossRef]
- Palma, M.S.; Itagaki, Y.; Fujita, T.; Naoki, H.; Nakajima, T. Structural characterization of a new acylpolyaminetoxin from the venom of Brazilian garden spider nephilengys cruentata. Toxicon 1998, 36, 485–493. [Google Scholar] [CrossRef]
- Aird, S.D.; Briones, A.V.; Roy, M.C.; Mikheyev, A.S. Polyamines as snake toxins and their probable pharmacological functions in envenomation. Toxins 2016, 8. [Google Scholar] [CrossRef] [PubMed]
- Hutchinson, D.A.; Savitzky, A.H.; Burghardt, G.M.; Nguyen, C.; Meinwald, J.; Schroeder, F.C.; Mori, A. Chemical defense of an Asian snake reflects local availability of toxic prey and hatchling diet. J. Zool. 2013, 289, 270–278. [Google Scholar] [CrossRef] [PubMed]
- Lai, L.C.; Hua, K.H.; Yang, C.C.; Huang, R.N.; Wu, W.J. Secretion Profiles of Venom Alkaloids in Solenopsis geminata (Hymenoptera: Formicidae) in Taiwan. Environ. Entomol. 2009, 38, 879–884. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Fadamiro, H.Y. Re-investigation of venom chemistry of Solenopsis fire ants. I. Identification of novel alkaloids in S. richteri. Toxicon 2009, 53, 469–478. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Lu, Y.Y.; Hu, Q.B.; Fadamiro, H.Y. Similarity in venom alkaloid chemistry of alate queens of imported fire ants: Implication for hybridization between Solenopsis richteri and S. invicta in the Southern United States. Chem. Biodivers. 2012, 9, 702–713. [Google Scholar] [CrossRef] [PubMed]
- Torres, V.D.O.; Piva, R.C.; Antonialli Junior, W.F.; Cardoso, C.A.L. Free Amino Acids Analysis in the Venom of the Social Wasp Polistes lanio Under Different Forms of Preservation. Orbital Electron. J. Chem. 2018, 10, 1–8. [Google Scholar] [CrossRef]
- Drexler, D.M.; Reily, M.D.; Shipkova, P.A. Advances in mass spectrometry applied to pharmaceutical metabolomics. Anal. Bioanal. Chem. 2011, 399, 2645–2653. [Google Scholar] [CrossRef] [PubMed]
- Klupczyńska, A.; Dereziński, P.; Kokot, Z.J. Metabolomics in Medical Sciences—Trends, Challenges and Perspectives. Acta Pol. Pharm. 2015, 72, 629–641. [Google Scholar] [PubMed]
- Heuberger, A.L.; Robison, F.M.; Lyons, S.M.A.; Broeckling, C.D.; Prenni, J.E. Evaluating plant immunity using mass spectrometry-based metabolomics workflows. Front. Plant Sci. 2014, 5, 291. [Google Scholar] [CrossRef] [PubMed]
- Roberts, L.D.; Souza, A.L.; Gerszten, R.E.; Clish, C.B. Targeted metabolomics. Curr. Protoc. Mol. Biol. 2012, 30.2.1–30.2.24. [Google Scholar] [CrossRef] [PubMed]
- Moroni, F.; Cozzi, A.; Sili, M.; Mannaioni, G. Kynurenic acid: A metabolite with multiple actions and multiple targets in brain and periphery. J. Neural Transm. 2012, 119, 133–139. [Google Scholar] [CrossRef] [PubMed]
- Strømgaard, K.; Andersen, K.; Krogsgaard-Larsen, P.; Jaroszewski, J.W. Recent advances in the medicinal chemistry of polyamine toxins. Mini Rev. Med. Chem. 2001, 1, 317–338. [Google Scholar] [CrossRef] [PubMed]
- Qi, F.; Li, A.; Inagaki, Y.; Kokudo, N.; Tamura, S.; Nakata, M.; Tang, W. Antitumor activity of extracts and compounds from the skin of the toad Bufo bufo gargarizans Cantor. Int. Immunopharmacol. 2011, 11, 342–349. [Google Scholar] [CrossRef] [PubMed]
- Terness, P.; Navolan, D.; Dufter, C.; Kopp, B.; Opelz, G. The T-cell suppressive effect of bufadienolides: Structural requirements for their immunoregulatory activity. Int. Immunopharmacol. 2001, 1, 119–134. [Google Scholar] [CrossRef]
- Aird, S.D. Ophidian envenomation strategies and the role of purines. Toxicon 2002, 40, 335–393. [Google Scholar] [CrossRef]
- Wilson, D.; Boyle, G.M.; McIntyre, L.; Nolan, M.J.; Parsons, P.G.; Smith, J.J.; Tribolet, L.; Loukas, A.; Liddell, M.J.; Rash, L.D.; et al. The aromatic head group of spider toxin polyamines influences toxicity to cancer cells. Toxins 2017, 9. [Google Scholar] [CrossRef] [PubMed]
- Estrada, G.; Villegas, E.; Corzo, G. Spider venoms: A rich source of acylpolyamines and peptides as new leads for CNS drugs. Nat. Prod. Rep. 2007, 24, 145–161. [Google Scholar] [CrossRef] [PubMed]
- Hassani, O.; Mansuelle, P.; Cestèle, S.; Bourdeaux, M.; Rochat, H.; Sampieri, F. Role of lysine and tryptophan residues in the biological activity of toxin VII (Ts γ) from the scorpion Tityus serrulatus. Eur. J. Biochem. 1999, 260, 76–86. [Google Scholar] [CrossRef] [PubMed]
- Abe, T.; Hariya, Y.; Kawai, N.; Miwa, A. Comparative study of amino acid composition in an extract from hornet venom sacs: High content of neuroactive amino acids in Vespa. Toxicon 1989, 27, 683–688. [Google Scholar] [CrossRef]
- Weisel-Eichler, A.; Libersat, F. Venom effects on monoaminergic systems. J. Comp. Physiol. A Neuroethol. Sens. Neural Behav. Physiol. 2004, 190, 683–690. [Google Scholar] [CrossRef] [PubMed]
- Krenn, L.; Kopp, B. Bufadienolides from animal and plant sources. Phytochemistry 1998, 48, 1–29. [Google Scholar] [CrossRef]
- Zhang, H.; Su, Y.; Yang, F.; Zhao, Z.; Gao, X. Two new bufadienolides and one new pregnane from Helleborus thibetanus. Phytochem. Lett. 2014, 10, 164–167. [Google Scholar] [CrossRef]
- Kolodziejczyk-Czepas, J.; Stochmal, A. Bufadienolides of Kalanchoe species: An overview of chemical structure, biological activity and prospects for pharmacological use. Phytochem. Rev. 2017, 16, 1155–1171. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Yu, Z.; Yang, Z.J.; Zhu, G.; Fong, W. Comprehensive chemical analysis of Venenum Bufonis by using liquid chromatography/electrospray ionization tandem mass spectrometry. J. Pharm. Biomed. Anal. 2011, 56, 210–220. [Google Scholar] [CrossRef] [PubMed]
- Ma, H.; Zhang, J.; Jiang, J.; Zhou, J.; Xu, H.; Zhan, Z.; Wu, Q.; Duan, J. Bilirubin attenuates bufadienolide-induced ventricular arrhythmias and cardiac dysfunction in guinea-pigs by reducing elevated intracellular Na+levels. Cardiovasc. Toxicol. 2012, 12, 83–89. [Google Scholar] [CrossRef] [PubMed]
- Cunha-Filho, G.A.; Resck, I.S.; Cavalcanti, B.C.; Pessoa, C.Ó.; Moraes, M.O.; Ferreira, J.R.O.; Rodrigues, F.A.R.; dos Santos, M.L. Cytotoxic profile of natural and some modified bufadienolides from toad Rhinella schneideri parotoid gland secretion. Toxicon 2010, 56, 339–348. [Google Scholar] [CrossRef] [PubMed]
- Tempone, A.G.; Pimenta, D.C.; Lebrun, I.; Sartorelli, P.; Taniwaki, N.N.; de Andrade, H.F.; Antoniazzi, M.M.; Jared, C. Antileishmanial and antitrypanosomal activity of bufadienolides isolated from the toad Rhinella jimi parotoid macrogland secretion. Toxicon 2008, 52, 13–21. [Google Scholar] [CrossRef] [PubMed]
- Kamano, Y.; Satoh, N.; Nakayoshi, H.; Pettit, G.R.; Smith, C.R. Rhinovirus inhibition by bufadienolides. Chem. Pharm. Bull. 1988, 36, 326–332. [Google Scholar] [CrossRef] [PubMed]
- Cunha Filho, G.A.; Schwartz, C.A.; Resck, I.S.; Murta, M.M.; Lemos, S.S.; Castro, M.S.; Kyaw, C.; Pires, O.R.; Leite, J.R.S.; Bloch, C.; et al. Antimicrobial activity of the bufadienolides marinobufagin and telocinobufagin isolated as major components from skin secretion of the toad Bufo rubescens. Toxicon 2005, 45, 777–782. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Lu, G.; Wang, H.; Zhang, J.; Duan, J.; Ma, H.; Wu, Q. Molecular structure-affinity relationship of bufadienolides and human serum albumin in vitro and molecular docking analysis. PLoS ONE 2015, 10, e0126669. [Google Scholar] [CrossRef] [PubMed]
- Perera Córdova, W.H.; Leitão, S.G.; Cunha-Filho, G.; Bosch, R.A.; Alonso, I.P.; Pereda-Miranda, R.; Gervou, R.; Touza, N.A.; Quintas, L.E.M.; Noël, F. Bufadienolides from parotoid gland secretions of Cuban toad Peltophryne fustiger (Bufonidae): Inhibition of human kidney Na+/K+-ATPase activity. Toxicon 2016, 110, 27–34. [Google Scholar] [CrossRef] [PubMed]
- Yusuf, M.; Firdaus, A.R.R.; Supratman, U. Computational study of bufadienolides from Indonesia’s kalanchoe pinnata as Na+/K+-ATPase inhibitor for anticancer agent. J. Young Pharm. 2017, 9, 475–479. [Google Scholar] [CrossRef]
- Chen, K.K.; Kovaříková, A. Pharmacology and toxicology of toad venom. J. Pharm. Sci. 1967, 56, 1535–1541. [Google Scholar] [CrossRef] [PubMed]
- Fiehn, O. Metabolomics—The link between genotypes and phenotypes. Plant Mol. Biol. 2002, 48, 155–171. [Google Scholar] [CrossRef] [PubMed]
- Wishart, D.S.; Feunang, Y.D.; Marcu, A.; Guo, A.C.; Liang, K.; Vázquez-Fresno, R.; Sajed, T.; Johnson, D.; Li, C.; Karu, N.; et al. HMDB 4.0: The human metabolome database for 2018. Nucleic Acids Res. 2018, 46, D608–D617. [Google Scholar] [CrossRef] [PubMed]
- Yan, Z.; Fang, Q.; Wang, L.; Liu, J.; Zhu, Y.; Wang, F.; Li, F.; Werren, J.H.; Ye, G. Insights into the venom composition and evolution of an endoparasitoid wasp by combining proteomic and transcriptomic analyses. Sci. Rep. 2016, 6. [Google Scholar] [CrossRef] [PubMed]
- Kazuma, K.; Masuko, K.; Konno, K.; Inagaki, H. Combined venom gland transcriptomic and venom peptidomic analysis of the predatory ant Odontomachus monticola. Toxins 2017, 9. [Google Scholar] [CrossRef] [PubMed]
- Matysiak, J.; Hajduk, J.; Pietrzak, Ł.; Schmelzer, C.E.H.; Kokot, Z.J. Shotgun proteome analysis of honeybee venom using targeted enrichment strategies. Toxicon 2014, 90, 255–264. [Google Scholar] [CrossRef] [PubMed]
- Matysiak, J.; Hajduk, J.; Mayer, F.; Hebeler, R.; Kokot, Z.J. Hyphenated LC-MALDI-ToF/ToF and LC-ESI-QToF approach in proteomic characterization of honeybee venom. J. Pharm. Biomed. Anal. 2016, 121, 69–76. [Google Scholar] [CrossRef] [PubMed]
- Touchard, A.; Aili, S.R.; Fox, E.G.P.; Escoubas, P.; Orivel, J.; Nicholson, G.M.; Dejean, A. The biochemical toxin arsenal from ant venoms. Toxins 2016, 8. [Google Scholar] [CrossRef] [PubMed]
- Fox, E.G.P.; Pianaro, A.; Solis, D.R.; Delabie, J.H.C.; Vairo, B.C.; MacHado, E.D.A.; Bueno, O.C. Intraspecific and intracolonial variation in the profile of venom alkaloids and cuticular hydrocarbons of the fire ant Solenopsis saevissima smith (Hymenoptera: Formicidae). Psyche 2012. [Google Scholar] [CrossRef]
- Walker, A.A.; Weirauch, C.; Fry, B.G.; King, G.F. Venoms of heteropteran insects: A treasure trove of diverse pharmacological toolkits. Toxins 2016, 8, 43. [Google Scholar] [CrossRef] [PubMed]
- Silva-Cardoso, L.; Caccin, P.; Magnabosco, A.; Patron, M.; Targino, M.; Fuly, A.; Oliveira, G.A.; Pereira, M.H.; Maria das Graças, T.; Souza, A.S.; et al. Paralytic activity of lysophosphatidylcholine from saliva of the waterbug Belostoma anurum. J. Exp. Biol. 2010, 213, 3305–3310. [Google Scholar] [CrossRef] [PubMed]

| Venom/Poison Source | Metabolomic Strategy | Method | Example Metabolites | Reference |
|---|---|---|---|---|
| Spider | Untargeted | HPLC-MS; NMR | Organic acids, nucleosides, amines, amino acids | Schroeder et al. (2008) [27] |
| Spider | Untargeted | HPLC-MS; NMR | Acylpolyamines | Tzouros et al. (2004) Tzouros et al. (2013) Hisada et al. (1998) Palma et al. (1998) [28,29,30,31] |
| Snake | Targeted | HPLC; NMR | Steroids (bufadienolides) | Hutchinson et al. (2013) [33] |
| Snake | Targeted | UHPLC-MS | Polyamines | Aird et al. (2016) [32] |
| Scorpion | Untargeted | DI-MS | Amino acids, amines | Hu et al. (2013) [23] |
| Toad | Untargeted | HPLC; HPLC-MS | Amino acids, steroids (bufadienolides) | Gao et al. (2010) [21] |
| Toad | Untargeted | DI-MS | Amines, steroids (bufadienolides) | Hu et al. (2013) [23] |
| Toad | Untargeted | HPLC; HPLC-MS | Alkaloids, steroids (bufadienolides) | Sciani et al. (2013) [22] |
| Toad | Untargeted | UHPLC-MS | Steroids (bufadienolides) | Ma et al. (2016) [19] |
| Toad | Targeted | UHPLC-MS | Steroids (bufadienolides) | Zhou et al. (2015) Meng et al. (2016) [20,24] |
| Frog | Untargeted | HPLC-MS; NMR | Amino acid derivative (kynurenic acid) | Mariano et al. (2015) [26] |
| Frog | Untargeted | HPLC; NMR | Amino acids, sugars | Cavalante et al. (2017) [25] |
| Ant | Untargeted | GC-MS | Piperidine alkaloids | Lai et al. (2009) Chen et al. (2009) Chen et al. (2012) [34,35,36] |
| Wasp | Targeted | HPLC | Amino acids | [37] |
| Compound | Bufobufo gargarizans | Bufo marinus | Bufo viridis | Rhinella crucifer | Rhinella marina | Rhinella major | Rhaebo guttatus |
|---|---|---|---|---|---|---|---|
| Arenobufagin | + | + | |||||
| Bufotalin | + | ||||||
| Bufalin | + | + | + | + | + | ||
| Cinobufagin | + | ||||||
| Cinobufotalin | + | ||||||
| Dehydrobufotenine | + | + | + | + | |||
| Desacetylcinobufagin | + | + | |||||
| Gamabufotalin | + | + | |||||
| Hellebrigenin | + | + | + | + | |||
| Hellebrigenol-3-O-sulfate | + | ||||||
| Marinobufagin | + | + | + | + | + | + | + |
| N′-N′-dimethyl-serotonin(Bufotenin) | + | + | |||||
| Resibufogenin | + | + | + | ||||
| Telocinobufagin | + | + | + | + | + |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Klupczynska, A.; Pawlak, M.; Kokot, Z.J.; Matysiak, J. Application of Metabolomic Tools for Studying Low Molecular-Weight Fraction of Animal Venoms and Poisons. Toxins 2018, 10, 306. https://0-doi-org.brum.beds.ac.uk/10.3390/toxins10080306
Klupczynska A, Pawlak M, Kokot ZJ, Matysiak J. Application of Metabolomic Tools for Studying Low Molecular-Weight Fraction of Animal Venoms and Poisons. Toxins. 2018; 10(8):306. https://0-doi-org.brum.beds.ac.uk/10.3390/toxins10080306
Chicago/Turabian StyleKlupczynska, Agnieszka, Magdalena Pawlak, Zenon J. Kokot, and Jan Matysiak. 2018. "Application of Metabolomic Tools for Studying Low Molecular-Weight Fraction of Animal Venoms and Poisons" Toxins 10, no. 8: 306. https://0-doi-org.brum.beds.ac.uk/10.3390/toxins10080306






